 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.217940.2 | ||||||||
| Common Name | Csa_7G062840, LOC101203707 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1092aa MW: 121455 Da PI: 5.8021 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.7 | 4.2e-57 | 20 | 136 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
++ k+rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktvrE+hekLKvg+++vl+cyYah+een++fqr
Cucsa.217940.2 20 VEAKHRWLRPAEICEILRNYTKFRIASEPPDRPSSGSLFLFDRKVLRYFRKDGHKWRKKKDGKTVREAHEKLKVGSIDVLHCYYAHGEENENFQR 114
5669******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+Lee+l +iv+vhylevk
Cucsa.217940.2 115 RSYWMLEEHLMHIVFVHYLEVK 136
*******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.166 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-80 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.2E-50 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 7.0E-7 | 488 | 590 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 1.2E-5 | 504 | 577 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 2.02E-15 | 505 | 589 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 1.87E-15 | 689 | 799 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.831 | 692 | 770 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.40E-11 | 693 | 797 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 5.8E-14 | 694 | 801 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF52540 | 1.75E-8 | 913 | 964 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.014 | 913 | 935 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.352 | 914 | 943 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.8E-4 | 915 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0038 | 936 | 958 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 937 | 961 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.3E-4 | 939 | 958 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1092 aa Download sequence Send to blast |
MADRGSYGLA PRLDIEQLLV EAKHRWLRPA EICEILRNYT KFRIASEPPD RPSSGSLFLF 60 DRKVLRYFRK DGHKWRKKKD GKTVREAHEK LKVGSIDVLH CYYAHGEENE NFQRRSYWML 120 EEHLMHIVFV HYLEVKGNRT NVGAVVETDE VSTSSQKSRS SSYSSSHNQA ASENADSPSP 180 TSTLTSFCED ADNDTYQATS RFHSFPTSPK MGNGLLVNKP DAGQSNFYFP HSSSNNAEAW 240 STVPAVDYVT QVQKDGLGGN GGDTSMMGSQ KTLSSASWEE ILHQCTTGFQ TVPSHVLTSS 300 IEPLPSGIVF GQENSTPDKL LTSNSAIKED FGSALAMTSN WQVPFEDNTL SFSKEHVDHF 360 PDLYSVCDID SRLTAQKSHD ATFGRGHEMF CAHPGKQNEE ILPNLELQFK EGESYSTARL 420 SSDNDMSKEG TISYSLTLKQ SLMDGEESLK KVDSFSRWVS KELGEVDDLH MHPSSGLTWT 480 TVECGDMVDD SSLSPSISED QLFSITAFSP KWTVADLDTE VVVIGRFMGN NNGTNCHWSC 540 MFGEVEVPAE VLADGILCCH APPHSVGRVP FYVTCSNRVA CSEVREFDYL AGSAQDVNVT 600 DIYNAGATEE LRMHLRFERL LSLEPSDPSN DLSESALEKQ NLIRELITIK EEDDTYGEDP 660 NPQNDQIQHQ SKEFLFVKLM KEKLYSWLSH KVIEGGKGPN ILDSEGQGVI HLAAALGYDW 720 AIRPIVAAGV SINFRDINGW TALHWAALCG RELTVGTLIT LDASPGLMSD PSPEVPLGIV 780 PADLASINGH KGISGFLAEA ALTSYVSSIS MAETVQDGVS DASRTKAVQT VSERRATPVN 840 DGFMPGDLSL KDSLTAVCNA TQAAGRIYQI LRVQSFQRKK LSECGTDEFG SSDNSILSFM 900 KARARKSGLS NNPAHAAAVQ IQKKFRGWRM RKEFLLIRQR IVKIQAHVRG HQVRKQYKKI 960 VWSVGMIDKI ILRWRRKGSG LRGFRSDAVP KDPPALMAPP TKEDDYDFLK EGRRQTEERF 1020 QKALTRVKSM AQYPEGRDQY RRLLTVVQKC RETKGSAMVV TTTSEEVIEG DDMIDIDTLL 1080 DDDALMSMTF D* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
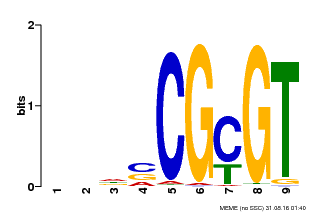
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681792 | 0.0 | LN681792.1 Cucumis melo genomic scaffold, anchoredscaffold00034. | |||
| GenBank | LN713255 | 0.0 | LN713255.1 Cucumis melo genomic chromosome, chr_1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011658786.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 isoform X3 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A0A0K2J6 | 0.0 | A0A0A0K2J6_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004137052.1 | 0.0 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.217940.2 |
| Entrez Gene | 101203707 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



