 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.272530.1 | ||||||||
| Common Name | Csa_4G664590, LOC101219733 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1032aa MW: 114643 Da PI: 8.5348 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.8 | 2.1e-40 | 109 | 186 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCe hsk++++lv++++qrfCqqCsrfh lsefD++krsCrrrLa+hn rrrk+q+
Cucsa.272530.1 109 MCQVDNCKEDLSNAKDYHRRHKVCELHSKSSKALVAKQMQRFCQQCSRFHPLSEFDDGKRSCRRRLAGHNWRRRKTQP 186
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.3E-33 | 106 | 171 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.317 | 107 | 184 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 8.76E-37 | 108 | 188 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.3E-29 | 110 | 183 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1032 aa Download sequence Send to blast |
MDDPGAQVVP PIFIHQSLTS RYTDLPSIPK KRPLSYHQGQ LHPHTWNPKA WDWDSSKFLT 60 KPSNLNNTTL DDHDDTLRLN LGGRYVEDPV SKPPKKVRPG SPASVTYPMC QVDNCKEDLS 120 NAKDYHRRHK VCELHSKSSK ALVAKQMQRF CQQCSRFHPL SEFDDGKRSC RRRLAGHNWR 180 RRKTQPEDVT SRLTRPGSRG PPSTGNLDIV SLLTVLARAQ GKNEDQSVKS LLSANSDQLI 240 QILNKINSLP LPADLAAKLP NLENFKGKAP PQSSLQHQNK LNGNPSSPST MDLLTVLSAT 300 LAASAPDALA MLSQKSSVSS DSEKTRSSCP SGSDLQNRPL ELPSVGGERS STSYQSPMED 360 SDGQVQGTRV GLPLQLFGSS PEHDAPPNLT ASRKYFSSDS SNPIEERSPS SSPPLLQTLF 420 PVQSTEETTS NGKMPIRKEV NGVEVRKPPS SNIPFELFRE LDGARPNSFQ TIHYQAGYTS 480 SGSDHSPSSL NSDAQDRTGR ISFKLFEKDP SQFPGTLRTQ IYNWLSNCPS EMESYIRPGC 540 VVLSVYMSMS SIAWERLEEN LVLHLKSLVH SEELDFWRSG RFLVYTGRQL ASHKDGKIHL 600 NKSSKAWSNP ELTSVSPLAV VSGQKTSFLL RGRNLKIPGT RIHCTSMGGY ISEEVMGLSS 660 LGLSSEGIYD EIHSRSFKVG DVSPTTLGRC FIEVENGFRG NSFPVIIADA TICRELRHLE 720 SDFDEFKVPD SSLESHSSVS SQPRLRDEIL QFLNELGWLF QRERFSYELD NPDFLIRRFR 780 FLLTFSAERD FCALVKTLLD ILAKKCLITD GLSMKSLEMI SELQLLNRSV KRRCRQMVDL 840 LVHYHVSGVG DSEKKYLFPP NFIGPGGITP LHLAASMADA ENLVDALTND PLEIGLECWS 900 SQLDESGRSP QAYALMRGNH NCNELVKRKL ADRKNGQVSV RIGNEIEQLE VSSGERGRVK 960 GRSCSRCAVV AARCNRRVPG SGTHRLLHRP YIHSMLAIAA VCVCVCLFLR GSPDIGLVAP 1020 FKWENLGYGT I* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-30 | 100 | 183 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
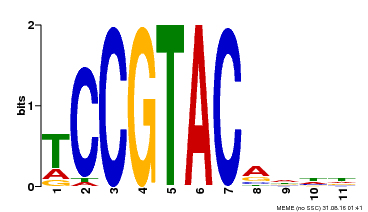
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681861 | 0.0 | LN681861.1 Cucumis melo genomic scaffold, anchoredscaffold00029. | |||
| GenBank | LN713261 | 0.0 | LN713261.1 Cucumis melo genomic chromosome, chr_7. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004145609.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A0A0L4Q1 | 0.0 | A0A0A0L4Q1_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004172805.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9694 | 31 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.272530.1 |
| Entrez Gene | 101219733 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



