 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.291770.1 | ||||||||
| Common Name | Csa_1G690370, LOC101211010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 619aa MW: 71109.5 Da PI: 6.5092 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 44.7 | 3.5e-14 | 124 | 186 | 1 | 68 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikege 68
+W+++e laL+++r+++e+ + ++ We+vs+k+ e gf+r++ +Ckek+e+ ++++ i+ ++
Cucsa.291770.1 124 QWSNDELLALLRIRSNIENCFPES-----TWEHVSRKLGEVGFRRTADKCKEKFEEESRYFNHINYNK 186
7******************99998.....9*******************************9988655 PP
| |||||||
| 2 | trihelix | 95.4 | 5.4e-30 | 487 | 582 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgk...........lkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdq 84
rW+++evlaL+++r +m ++ +++ lk+plWe++s+ m + g++rs+k+Ckekwen+nk+++k+k+ +kkr s +s+tcpyf+q
Cucsa.291770.1 487 RWPRDEVLALVNVRCKMYNNTTTTNnqdesqsggasLKAPLWERISQGMLQLGYKRSAKRCKEKWENINKYFRKTKDVNKKR-SLDSRTCPYFHQ 580
8**************888888876445566666677*********************************************8.9999******** PP
trihelix 85 le 86
l
Cucsa.291770.1 581 LS 582
95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 22 | 121 | 178 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 7.8E-9 | 123 | 184 | No hit | No description |
| PROSITE profile | PS50090 | 6.481 | 125 | 176 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 0.52 | 331 | 557 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 4.6E-18 | 486 | 582 | No hit | No description |
| CDD | cd12203 | 2.41E-22 | 486 | 562 | No hit | No description |
| PROSITE profile | PS50090 | 6.179 | 487 | 555 | IPR017877 | Myb-like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001158 | Molecular Function | enhancer sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 619 aa Download sequence Send to blast |
MFEGGSVSEQ LHQFLTPRTT PPPPNSNSLP LIPLNFALHS PNFNFHPFDS YNATSTAHHH 60 HQIHLPHHLL HHQSPNPHGD DKNDVKTTTT AAGSSLQVGV DLEVGRENSR SILMEDHHII 120 HHDQWSNDEL LALLRIRSNI ENCFPESTWE HVSRKLGEVG FRRTADKCKE KFEEESRYFN 180 HINYNKNCRF LTHELNYNHH PNQDQDQDHL LLIHEGNGKP DDGGPTLVVV PEEGEEENQD 240 KDGELHDDDE EEDLRNDEMR PGRNEEERNE SSRSSSCQKS KKKRKMMRQK EFELLKGYCE 300 EIVKKMMIQQ EEIHSKLLQD MLKKEEEKVA KEEYWKKEQM ERLHKELEVM AHEQAIAGDR 360 QATIIEILNQ ITNSTTLISS SHESKKDLQN LLQSLNNYNN NNNIPNSTPS SSSLIQCQTS 420 SSPNKKPPHE NSNSFTSQND PIKNPKNNPC LSTQILAPQD PNSFINNHPN PKSKEKLDHE 480 SEDLGKRWPR DEVLALVNVR CKMYNNTTTT NNQDESQSGG ASLKAPLWER ISQGMLQLGY 540 KRSAKRCKEK WENINKYFRK TKDVNKKRSL DSRTCPYFHQ LSTLYNQGGG NNNPLENCPN 600 VSSENHSDHS ENHLATSS* |
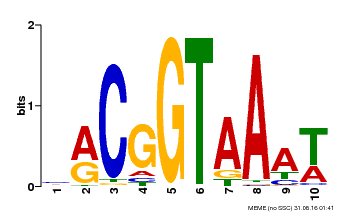
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00011 | PBM | Transfer from AT5G28300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681811 | 0.0 | LN681811.1 Cucumis melo genomic scaffold, anchoredscaffold00069. | |||
| GenBank | LN713256 | 0.0 | LN713256.1 Cucumis melo genomic chromosome, chr_2. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142523.1 | 0.0 | PREDICTED: trihelix transcription factor GTL2 | ||||
| TrEMBL | A0A0A0M3J3 | 0.0 | A0A0A0M3J3_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004142523.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004155698.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5043 | 33 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G28300.1 | 4e-61 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.291770.1 |
| Entrez Gene | 101211010 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



