 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.302100.2 | ||||||||
| Common Name | Csa_5G160210, LOC101214748 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | YABBY | ||||||||
| Protein Properties | Length: 133aa MW: 14932.8 Da PI: 10.3907 | ||||||||
| Description | YABBY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | YABBY | 138.5 | 7.8e-43 | 24 | 92 | 102 | 170 |
YABBY 102 senedeevprvppvirPPekrqrvPsaynrfikeeiqrikasnPdishreafsaaaknWahfPkihfgl 170
s+ ++e+pr+p v+rPPekrqrvPsaynrfik+eiqrik+ nPdishreafsaaaknWahfP+ihfgl
Cucsa.302100.2 24 SRGVTDELPRPPVVNRPPEKRQRVPSAYNRFIKDEIQRIKSVNPDISHREAFSAAAKNWAHFPHIHFGL 92
6778899***999******************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04690 | 1.1E-38 | 31 | 92 | IPR006780 | YABBY protein |
| SuperFamily | SSF47095 | 3.93E-8 | 36 | 85 | IPR009071 | High mobility group box domain |
| Gene3D | G3DSA:1.10.30.10 | 9.2E-5 | 40 | 86 | IPR009071 | High mobility group box domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009909 | Biological Process | regulation of flower development | ||||
| GO:0009933 | Biological Process | meristem structural organization | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0010093 | Biological Process | specification of floral organ identity | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010158 | Biological Process | abaxial cell fate specification | ||||
| GO:0010159 | Biological Process | specification of organ position | ||||
| GO:0010450 | Biological Process | inflorescence meristem growth | ||||
| GO:1902183 | Biological Process | regulation of shoot apical meristem development | ||||
| GO:2000024 | Biological Process | regulation of leaf development | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 133 aa Download sequence Send to blast |
MATPNSNYLI NQFGATTNNF GMPSRGVTDE LPRPPVVNRP PEKRQRVPSA YNRFIKDEIQ 60 RIKSVNPDIS HREAFSAAAK NWAHFPHIHF GLMPDQTVKK TNIRQQEQGD AQNVLMKDNG 120 FYASANVGVT HF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the abaxial cell fate determination during embryogenesis and organogenesis. Regulates the initiation of embryonic shoot apical meristem (SAM) development (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6, PubMed:19837869). Required during flower formation and development, particularly for the patterning of floral organs. Positive regulator of class B (AP3 and PI) activity in whorls 2 and 3. Negative regulator of class B activity in whorl 1 and of SUP activity in whorl 3. Interacts with class A proteins (AP1, AP2 and LUG) to repress class C (AG) activity in whorls 1 and 2. Contributes to the repression of KNOX genes (STM, KNAT1/BP and KNAT2) to avoid ectopic meristems. Binds DNA without sequence specificity. In vitro, can compete and displace the AP1 protein binding to DNA containing CArG box (PubMed:10323860, PubMed:10331982, PubMed:10457020, PubMed:11812777, PubMed:12417699, PubMed:9878633, Ref.3, Ref.6). {ECO:0000269|PubMed:10323860, ECO:0000269|PubMed:10331982, ECO:0000269|PubMed:10457020, ECO:0000269|PubMed:11812777, ECO:0000269|PubMed:12417699, ECO:0000269|PubMed:19837869, ECO:0000269|PubMed:9878633, ECO:0000269|Ref.3, ECO:0000269|Ref.6}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00620 | PBM | Transfer from AT2G45190 | Download |
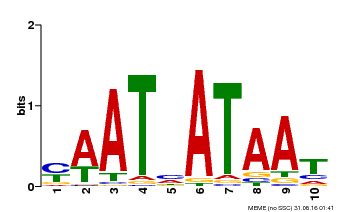
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN683796 | 5e-49 | LN683796.1 Cucumis melo genomic scaffold, unassembled_sequence31283. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004147586.1 | 6e-96 | PREDICTED: axial regulator YABBY 5-like | ||||
| Swissprot | O22152 | 3e-47 | YAB1_ARATH; Axial regulator YABBY 1 | ||||
| TrEMBL | A0A0A0KKT0 | 1e-94 | A0A0A0KKT0_CUCSA; Axial regulator YABBY1 | ||||
| STRING | XP_004147586.1 | 2e-95 | (Cucumis sativus) | ||||
| STRING | XP_004167326.1 | 2e-95 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45190.1 | 1e-49 | YABBY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.302100.2 |
| Entrez Gene | 101214748 |



