 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.302120.1 | ||||||||
| Common Name | Csa_5G160180, LOC101221404 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 330aa MW: 37861.7 Da PI: 6.8775 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 180.6 | 3.9e-56 | 7 | 137 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk....kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdke 91
+ppGfrFhPtdeelv++yL+kkv++k+++l ++ik+vd+y++ePwdL++ + +++++ewyfFs++dkky+tg+r+nratk+g+Wkatg+dk+
Cucsa.302120.1 7 VPPGFRFHPTDEELVDYYLRKKVASKRIDL-DIIKDVDLYRIEPWDLQElcklAGSEDQNEWYFFSHKDKKYPTGTRTNRATKAGFWKATGRDKA 100
69****************************.99**************9545433333667*********************************** PP
NAM 92 vlskkgelvglkktLvfykgrapkgektdWvmheyrle 129
+++ +++lvg++ktLvfykgrap+g+k+dW+mheyrle
Cucsa.302120.1 101 IYA-RHSLVGMRKTLVFYKGRAPNGQKSDWIMHEYRLE 137
***.889*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 3.66E-61 | 5 | 157 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 60.017 | 7 | 157 | IPR003441 | NAC domain |
| Pfam | PF02365 | 6.4E-29 | 8 | 136 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
MNCIPHVPPG FRFHPTDEEL VDYYLRKKVA SKRIDLDIIK DVDLYRIEPW DLQELCKLAG 60 SEDQNEWYFF SHKDKKYPTG TRTNRATKAG FWKATGRDKA IYARHSLVGM RKTLVFYKGR 120 APNGQKSDWI MHEYRLETNE NATPQEEGWV VCRVFKKRMP TVRKAGDYGS PCWYDDQVSF 180 MPELDSPTTQ IFHHPAYPCK QELELHFHAP PHDQVFLQLP HLESPKIPTS SATNSLLPYA 240 YDRDTGVTPL YGVEHTVDHV TDWRVMEKFV ASQLSNDQDL KQSNYSHGAI FQVPPNSGGA 300 ATVSECDRKD MAPEFALASP SSCQVDMWK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 1e-48 | 6 | 158 | 14 | 169 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:16103214, PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
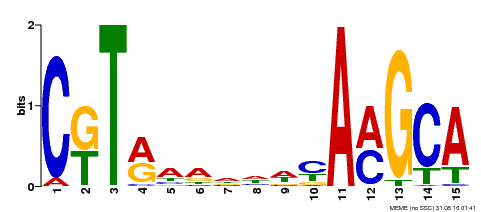
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00136 | DAP | Transfer from AT1G12260 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681895 | 0.0 | LN681895.1 Cucumis melo genomic scaffold, anchoredscaffold00005. | |||
| GenBank | LN713263 | 0.0 | LN713263.1 Cucumis melo genomic chromosome, chr_9. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004147614.1 | 0.0 | PREDICTED: NAC domain-containing protein 7 | ||||
| Swissprot | Q9FWX2 | 1e-138 | NAC7_ARATH; NAC domain-containing protein 7 | ||||
| TrEMBL | A0A0A0KKJ4 | 0.0 | A0A0A0KKJ4_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004147614.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004167328.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1269 | 33 | 106 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G12260.1 | 1e-141 | NAC 007 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.302120.1 |
| Entrez Gene | 101221404 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



