 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.307870.1 | ||||||||
| Common Name | Csa_5G623900 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 326aa MW: 37364.6 Da PI: 7.9267 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.1 | 7.5e-06 | 22 | 42 | 3 | 23 |
ETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 CpdCgksFsrksnLkrHirtH 23
C+ Cg F+ + +Lk+H+++H
Cucsa.307870.1 22 CQQCGAAFRKPAYLKQHMQSH 42
*******************99 PP
| |||||||
| 2 | zf-C2H2 | 16 | 3.5e-05 | 48 | 72 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y C+ C+ s++rk++L+rH+ H
Cucsa.307870.1 48 YICSvdGCNASYRRKDHLTRHLLHH 72
899999***************9987 PP
| |||||||
| 3 | zf-C2H2 | 18.2 | 6.9e-06 | 125 | 149 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp Cgk+F+ s L++H +H
Cucsa.307870.1 125 FVCPedGCGKVFRYASRLQKHEGSH 149
89*******************8666 PP
| |||||||
| 4 | zf-C2H2 | 23.8 | 1.1e-07 | 218 | 241 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirt.H 23
C+ C+k+Fs+ snL++H+++ H
Cucsa.307870.1 218 CTyeGCSKVFSTASNLQQHMKVvH 241
88889****************988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00355 | 0.017 | 20 | 42 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.048 | 20 | 47 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 22 | 42 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 8.4E-9 | 22 | 47 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 4.22E-11 | 34 | 74 | No hit | No description |
| SMART | SM00355 | 0.0041 | 48 | 72 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.2E-10 | 48 | 74 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.494 | 48 | 77 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 50 | 72 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.6 | 77 | 102 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.759 | 77 | 107 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 79 | 102 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.4E-8 | 119 | 149 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.32E-5 | 122 | 150 | No hit | No description |
| PROSITE profile | PS50157 | 10.762 | 125 | 154 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 6.8E-4 | 125 | 149 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 127 | 149 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.55 | 157 | 182 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.7E-4 | 159 | 182 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 159 | 182 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.6 | 185 | 207 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.7E-8 | 197 | 253 | No hit | No description |
| SMART | SM00355 | 7.6E-5 | 216 | 241 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.006 | 216 | 246 | IPR007087 | Zinc finger, C2H2 |
| Pfam | PF00096 | 2.4E-5 | 217 | 241 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.9E-8 | 218 | 241 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 218 | 241 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-4 | 242 | 269 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.8 | 247 | 273 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 249 | 273 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MEIGKHNDDG DDKTKSGKFN ICQQCGAAFR KPAYLKQHMQ SHSLERPYIC SVDGCNASYR 60 RKDHLTRHLL HHQGTMFKCP IESCCREFAY ASCVSRHLKE CHEGEVPPEE CHEGEAPPEE 120 CKKQFVCPED GCGKVFRYAS RLQKHEGSHV KLDSIEACCT EPGCLKTFTN KQCLRAHMQT 180 CHQYVKCEIC GEKKLRKNLK QHLRQKHESE GPLEAIPCTY EGCSKVFSTA SNLQQHMKVV 240 HLEQKPFVCC FPGCGMRFGY KHVRDNHEKS GQHVYTNGDF EAADEQFRSK PRGGRKRICP 300 TVEMLIRKRV TPPSELDFMM EEEDY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5v3m_C | 3e-15 | 22 | 281 | 64 | 314 | Zinc finger protein 568 |
| 5wjq_D | 3e-15 | 22 | 277 | 39 | 285 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
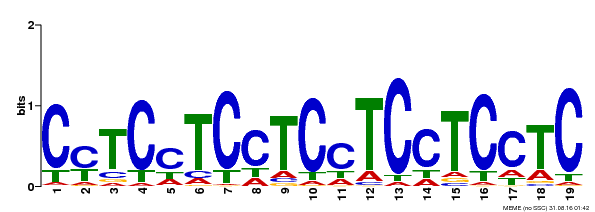
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU284120 | 0.0 | EU284120.1 Cucumis sativus C2H2 zinc finger protein 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142387.2 | 0.0 | PREDICTED: zinc finger X-linked protein ZXDB | ||||
| Swissprot | Q84MZ4 | 1e-119 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0A0KV54 | 0.0 | A0A0A0KV54_CUCSA; Uncharacterized protein | ||||
| STRING | XP_008446968.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3069 | 33 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 4e-86 | transcription factor IIIA | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.307870.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



