 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.309430.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 220aa MW: 24723.8 Da PI: 8.5637 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 109.8 | 2.1e-34 | 14 | 102 | 1 | 89 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleq 89
aCaaCk++r+kC + C+lapyfpae++++f++vhk+FG+snv+k++k+++ee++++a++sl++eA +r++dPv G +g ++++ ++++
Cucsa.309430.1 14 ACAACKHQRKKCHETCPLAPYFPAERNREFQAVHKVFGVSNVTKMVKNVREEDKRKAVDSLIWEAVCRQNDPVLGPYGEYKRVLEEVKV 102
7*****************************************************************************99998887765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 22.838 | 13 | 113 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 2.6E-33 | 14 | 102 | IPR004883 | Lateral organ boundaries, LOB |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 220 aa Download sequence Send to blast |
MQKHNNGVLQ GTPACAACKH QRKKCHETCP LAPYFPAERN REFQAVHKVF GVSNVTKMVK 60 NVREEDKRKA VDSLIWEAVC RQNDPVLGPY GEYKRVLEEV KVCKLLTQNQ SIMNGGQVGL 120 GYKPIIPCLG AWNNNNNNNS NDNNNNNGNG NGVNSSHLLN YFHDNYVSAI VDSSPYSNSS 180 AYNLQSHEKL MRLQQDKEIS SNMVVPLQQQ QHPTFHHCF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 3e-18 | 11 | 91 | 9 | 88 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 3e-18 | 11 | 91 | 9 | 88 | LOB family transfactor Ramosa2.1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00128 | DAP | Transfer from AT1G06280 | Download |
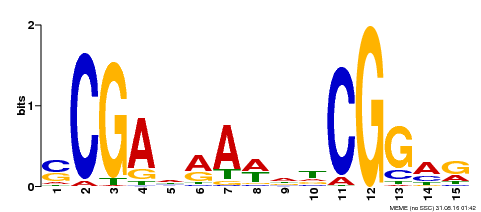
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681916 | 0.0 | LN681916.1 Cucumis melo genomic scaffold, anchoredscaffold00037. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016901865.1 | 1e-128 | PREDICTED: LOB domain-containing protein 2 | ||||
| TrEMBL | A0A1S4E1K1 | 1e-127 | A0A1S4E1K1_CUCME; LOB domain-containing protein 2 | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF12722 | 25 | 34 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06280.1 | 3e-45 | LOB domain-containing protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.309430.1 |



