 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.325250.1 | ||||||||
| Common Name | Csa_6G309970, LOC101211298 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 917aa MW: 104047 Da PI: 7.7439 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 165.1 | 1.2e-51 | 35 | 150 | 4 | 118 |
CG-1 4 e.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
e ++rwl+++ei+aiL n++ ++++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ve +++yYah+ + ptf rr
Cucsa.325250.1 35 EaSARWLRPNEIHAILCNYKYFTIHVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNVERIHVYYAHGLDSPTFVRR 129
4589******************************************************************************************* PP
CG-1 98 cywlLeeelekivlvhylevk 118
cywlL+++le+ivlvhy+e++
Cucsa.325250.1 130 CYWLLDKTLEHIVLVHYRETQ 150
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.854 | 29 | 155 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 7.0E-73 | 32 | 150 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.4E-45 | 35 | 148 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.4E-8 | 358 | 458 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 2.24E-17 | 369 | 456 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 1.2E-8 | 370 | 455 | IPR002909 | IPT domain |
| CDD | cd00102 | 1.15E-4 | 370 | 456 | No hit | No description |
| CDD | cd00204 | 1.98E-16 | 531 | 665 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 5.9E-17 | 555 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 5.44E-19 | 563 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 16.016 | 573 | 638 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 5.9E-7 | 578 | 668 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 8.9E-6 | 606 | 635 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.487 | 606 | 638 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 3100 | 645 | 674 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 9.96E-6 | 715 | 820 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 6.577 | 717 | 746 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 64 | 769 | 791 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.199 | 770 | 797 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.028 | 792 | 814 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 793 | 817 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.016 | 795 | 814 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 9.5 | 870 | 892 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 872 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 917 aa Download sequence Send to blast |
MSLSMKADVQ GSLVGSEIHG FHTLQDLDVE NIREEASARW LRPNEIHAIL CNYKYFTIHV 60 KPVNLPKSGT IVLFDRKMLR NFRKDGHNWK KKKDGKTVKE AHEHLKVGNV ERIHVYYAHG 120 LDSPTFVRRC YWLLDKTLEH IVLVHYRETQ EFQNSPSTSL NSNSGSVSNP STPWLLSEEL 180 DSKATHVYSV GENELSEPSD TTTVMTHEQR LHEINTLEWD DLLVKDEPFK PAIHKGDKLS 240 CFDQQNQVPI NATSNLLGEM SSFSNPVEST GRANGNISFT GSANLLLGGQ TNLNVEKRES 300 IAINSTDNLL DERLQSQDSF GRWINEVIIE SPGSVIDPAI EPSISYVHNS YRDSTLYHSQ 360 TLATEQIFNI TDVSPSWAFS TEKTKILIIG YFHNDFVHLA KSNLLVVCGD TSVNVDFVQP 420 GVYRCLVPPH APGLVHLYVS VDGHKPISQA LNFEYRAPNL EVPVVASEQS QKWEEFQIQM 480 RLAHMLFSTS KILSIISTKL LPTALQEAKK LAVKTADISD SWIYLLKSIT ENRTPFQQAR 540 EGVLEIVLRS RLREWLIERV AEGAKKSTEF DVNGQGVIHL CAILGYTWAV HLFDWAGLSI 600 NFRDKFGWTA LHWAAYYGRE RMVAVLLSAG AKPNLVTDPS SKNPLGCTAA DLASMNGYDG 660 LAAYLSEKAL VSHFKEMSLA GNVSGSLDTS STITDTSDCI SEEQMYMKET LAAYRTAADA 720 ASRIQAAFRE HSLKQRSDRI ELSSPEDEAR SIIAAMKIQH AYRNFETRKK MAAAARIQYR 780 FRTWKIRKDF LNMRRQTIRI QAAFRGFQVR RQYRKIVWSV GVLEKAILRW RLKRKGFRGL 840 QVAPTEMVEK QQSDVEEDFY LVSQKQAEER VERAVVRVQA MFRSKKAQEE YRRMRLTCDE 900 AALEYEVLSH PVYGND* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
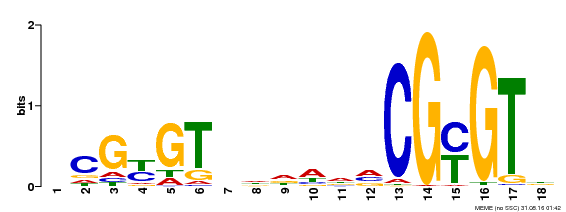
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681924 | 0.0 | LN681924.1 Cucumis melo genomic scaffold, anchoredscaffold00047. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011657270.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A0A0KCD9 | 0.0 | A0A0A0KCD9_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004161401.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.325250.1 |
| Entrez Gene | 101211298 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



