 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.328060.1 | ||||||||
| Common Name | Csa_6G373190, LOC101217750 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 333aa MW: 36517.2 Da PI: 6.0055 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.1 | 4.9e-16 | 51 | 100 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h+ E+rRR++iN++f+ Lr+l+P++ ++K + a+ L +++eY+++Lq
Cucsa.328060.1 51 RSKHSVTEQRRRSKINERFQILRDLIPHS----DQKRDTASFLLEVIEYVQYLQ 100
899*************************9....8*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 9.0E-23 | 47 | 120 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.49E-18 | 47 | 119 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.044 | 49 | 99 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.41E-15 | 51 | 104 | No hit | No description |
| Pfam | PF00010 | 2.3E-13 | 51 | 100 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.6E-12 | 55 | 105 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MRTGKSNQED DEYDEDELVG KREGPSSTSN AISSIGNSKD AKNNDKANAI RSKHSVTEQR 60 RRSKINERFQ ILRDLIPHSD QKRDTASFLL EVIEYVQYLQ EKVQKYEGSY QNWSGEPTKL 120 IPWRNSHWRM QTFVGHQQSV KNGSASGPTY SGKFDDNNIS ISPAMLASSQ NPLDSHSGRD 180 ILQEADITNK VPHPMSLEGN MHASIRSDTV LDHSLHIPIS DPQATECPIT NNASNQPEEM 240 GIEGGTISVS SVYSEGFMNS LAQALQSTGL DLSQASISVQ IDLGKRANKG MPFGTPIFKE 300 TGNPSSNHPA MANVGDLGSG DDSEQAKKRL RT* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
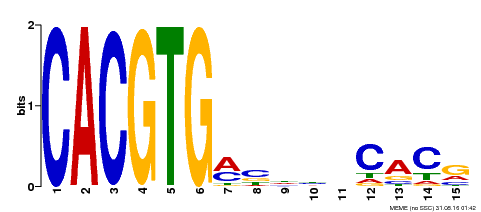
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by heat treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681823 | 0.0 | LN681823.1 Cucumis melo genomic scaffold, anchoredscaffold00014. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011657414.1 | 0.0 | PREDICTED: transcription factor BIM2 | ||||
| Swissprot | Q9CAA4 | 1e-105 | BIM2_ARATH; Transcription factor BIM2 | ||||
| TrEMBL | A0A0A0KD98 | 0.0 | A0A0A0KD98_CUCSA; Uncharacterized protein | ||||
| STRING | XP_008445413.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1730 | 33 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 1e-100 | BES1-interacting Myc-like protein 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.328060.1 |
| Entrez Gene | 101217750 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



