 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.342870.1 | ||||||||
| Common Name | Csa_3G866510, LOC101216825 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 968aa MW: 106722 Da PI: 5.0526 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 76.8 | 2.4e-24 | 149 | 250 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrs 88
f+k+lt sd++++g +++p++ ae+ + ++++ ++l+++d++ ++W++++iyr++++r++lt+GW+ Fv a++L++gD+v+F +++
Cucsa.342870.1 149 FCKTLTASDTSTHGGFSVPRRAAEKLfppldytM-QPPT-QELIVRDLHDNTWTFRHIYRGQPKRHLLTTGWSLFVGAKRLRAGDSVLFI--RDE 239
99*********************999*****954.4444.38************************************************..457 PP
EE..EEEEE-S CS
B3 89 efelvvkvfrk 99
+ +l+++v+r+
Cucsa.342870.1 240 KSQLLIGVRRA 250
7778****997 PP
| |||||||
| 2 | Auxin_resp | 113.5 | 2e-37 | 275 | 358 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+++s+F+++YnPra++seFv++++k++k ++ +++s+GmRf m+fete+s +rr++Gt+vg+sdldp+rWp+SkWr+L+
Cucsa.342870.1 275 AAHAAANRSPFTIFYNPRACPSEFVIPLAKYRKCVYgTQLSAGMRFGMMFETEESGKRRYMGTIVGISDLDPLRWPGSKWRNLQ 358
79*********************************99*********************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.05E-46 | 136 | 278 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.5E-42 | 142 | 263 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.35E-22 | 148 | 249 | No hit | No description |
| Pfam | PF02362 | 1.1E-21 | 149 | 250 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.7E-24 | 149 | 251 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.084 | 149 | 251 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.9E-32 | 275 | 358 | IPR010525 | Auxin response factor |
| PROSITE profile | PS51745 | 21.836 | 853 | 937 | IPR000270 | PB1 domain |
| Pfam | PF02309 | 8.6E-9 | 864 | 940 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 9.81E-9 | 868 | 932 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0009942 | Biological Process | longitudinal axis specification | ||||
| GO:0010305 | Biological Process | leaf vascular tissue pattern formation | ||||
| GO:0048364 | Biological Process | root development | ||||
| GO:0048507 | Biological Process | meristem development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016020 | Cellular Component | membrane | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 968 aa Download sequence Send to blast |
MGSVEEKLKT SGGLINNAPQ TNLLDEMKLL KEMQDQSGAR KAINSELWHA CAGPLVSLPH 60 VGSLVYYFPQ GHSEQVAVST KRTATSQIPN YPNLPSQLMC QVQNVTLHAD KDSDEIYAQM 120 SLQPVNSEKD VFLVPDFGLR PSKHPNEFFC KTLTASDTST HGGFSVPRRA AEKLFPPLDY 180 TMQPPTQELI VRDLHDNTWT FRHIYRGQPK RHLLTTGWSL FVGAKRLRAG DSVLFIRDEK 240 SQLLIGVRRA NRQQTTLPSS VLSADSMHIG VLAAAAHAAA NRSPFTIFYN PRACPSEFVI 300 PLAKYRKCVY GTQLSAGMRF GMMFETEESG KRRYMGTIVG ISDLDPLRWP GSKWRNLQVE 360 WDEPGCCDKQ NRVSSWEIET PESLFIFPSL TSGLKRPLHG GFLAGETDWG SLVKRPMLRV 420 PENIRGDLSY APTLCSEPLM KMLLRPQMVN LNGTTLQQDS TNNLVKIQDM KDMQNPKMQQ 480 LIPTETASPG NQNQHHPGPA QSDPINPNSS PKANVPGKVQ TSVAIESEAP TAADGDKAKY 540 DRDLSASTNQ SNPLPPVGGC AEEKLTSNEM NMQTLVNQLS FVNQNQIPMQ LQSVSWPMQP 600 QLESLIQHPQ PIDMPQPEYT NSNGLISSLD GDGCLINPSC LPLPGVMRSP GNLSMLGLQD 660 SSTVFPEVLN FPLPSTGQDM WDPLNNIRFS SQTNHLISFS HADASNLNCM ANANIMRDVS 720 DESNNQSGIY SCSNLEMSNG GSTLVDHAVS STILDDYCTL KDADFPHPSD CLAGNFSSSQ 780 DVQSQITSAS LGDSQAFSRQ EFHDNSAGTS SCNVDFDEGS LLQNGSWKQV VPPLRTYTKV 840 GTCDFYFCYF LELKDTEVQK AGSVGRSIDV TSFKNYDELC SAIECMFGLE GLLNDPRGSG 900 WKLVYVDYEN DVLLIGDDPW EEFVSCVRCI RILSPSEVQQ MSEEGMKLLN SAMMQGINCP 960 MSEGGRS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 0.0 | 1 | 383 | 5 | 392 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). Seems to act as transcriptional activator. Formation of heterodimers with Aux/IAA proteins may alter their ability to modulate early auxin response genes expression. Mediates embryo axis formation and vascular tissues differentiation. Functionally redundant with ARF7. May be necessary to counteract AMP1 activity. {ECO:0000269|PubMed:12036261, ECO:0000269|PubMed:14973283, ECO:0000269|PubMed:17553903}. | |||||
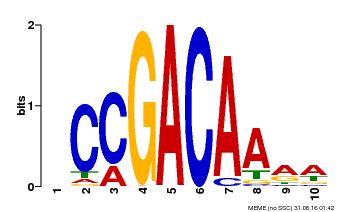
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00153 | DAP | Transfer from AT1G19850 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KU323419 | 0.0 | KU323419.1 UNVERIFIED: Trichosanthes dioica clone TdARF_27 auxin response factor 5-like mRNA, partial sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004147836.1 | 0.0 | PREDICTED: auxin response factor 5 isoform X1 | ||||
| Swissprot | P93024 | 0.0 | ARFE_ARATH; Auxin response factor 5 | ||||
| TrEMBL | A0A0A0LE69 | 0.0 | A0A0A0LE69_CUCSA; Auxin response factor | ||||
| STRING | XP_004154860.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF8991 | 32 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19850.1 | 0.0 | ARF family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.342870.1 |
| Entrez Gene | 101216825 |



