 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.391680.1 | ||||||||
| Common Name | Csa_1G050050 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1552aa MW: 173969 Da PI: 8.5383 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 11.8 | 0.00076 | 1518 | 1544 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
Cucsa.391680.1 1518 YVCAepGCGQTFRFVSDFSRHKRKtgH 1544
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 5.5E-17 | 23 | 64 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.284 | 24 | 65 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 1.2E-13 | 25 | 58 | IPR003349 | JmjN domain |
| SMART | SM00558 | 7.8E-48 | 196 | 365 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 32.261 | 199 | 365 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 3.3E-25 | 210 | 378 | No hit | No description |
| Pfam | PF02373 | 2.0E-36 | 229 | 348 | IPR003347 | JmjC domain |
| Gene3D | G3DSA:3.30.160.60 | 2.5E-4 | 1427 | 1457 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 17 | 1435 | 1457 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.136 | 1458 | 1487 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.072 | 1458 | 1482 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 3.8E-4 | 1459 | 1486 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1460 | 1482 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.55E-13 | 1469 | 1523 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.6E-8 | 1487 | 1512 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1488 | 1512 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.741 | 1488 | 1517 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1490 | 1512 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 2.3E-9 | 1513 | 1541 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.032 | 1518 | 1549 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.62 | 1518 | 1544 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1520 | 1544 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1552 aa Download sequence Send to blast |
MAGTAMAAEP TQEVLSWLKT LPLAPEYHPT LAEFQDPISY IFKIEKEASK FGICKIVPPV 60 PPSPKKTVIV NFNKSLAARA APCSDSTNSK SPPTFTTRQQ QIGFCPRKTR PVQKSVWQSG 120 EYYTFQQFEA KAKNFEKSYL KKCTKKGGLS PLEIETLYWR ATLDKPFSVE YANDMPGSAF 180 VPVSAKMFRE AGEGTTLGET AWNMRGVSRA KGSLLKFMKE EIPGVTSPMV YVAMMFSWFA 240 WHVEDHDLHS LNYLHMGAGK TWYGVPRDAA VAFEEVVRVQ GYGGEINPLV TFAVLGEKTT 300 VMSPEVLVSA GVPCCRLVQN AGEFVVTFPR AYHTGFSHGF NCGEAANIAT PEWLNVAKDA 360 AIRRASINYP PMVSHYQLLY DLALSSRAPL CTGAEPRSSR LKDKRRSEGD TVIKELFVQN 420 IVENNSLLDN LGGGASVVLL PPGSLESIYS RLRVGSHLRS KPRFPTGVCS SKEETKSPQS 480 FDYDNLALEN SPVINRVKGF YSANGPYSTL SERSTDNVCA SSLRPLNANN ERGGNVQSNG 540 LSDQRLFSCV TCGILSFACV AIIQPREQAA RYLMSADCSF FNDWVVGSGI ASEGISTRDR 600 HPVSSQQISN SGKRDKCVSD GLYDVPVQAV NRQLPLAGES YEANLNTEKR NETSALGMLA 660 LTYGHSSDSE EDNAEADAAL NVDDAKLMIC SSEDQYQFEN SGLTSGEYSK NTAILNHDPS 720 SFGINSADHM QFQVNDYEEF RRADSKDSFN CSSESEMDGI GSTKKNGLTR YQDSHVNGRS 780 SLDADTEKPV FDKSTETVET ENMPFAPDID EDFSRLHVFC LEHAKEVEQQ LRPIGGVHIL 840 LLCHPDYPKM EAEAKLVAQE LSMSHLWTDT IFRDATQDEE KRIQLALDSE EAIPGNGDWA 900 VKLGINLFYS ANLSHSPLYS KQMPYNSVIY NAFGRSTSAN SSGKPKVYQR RTGKLKRVVA 960 GKWCGKVWMS NQVHPLLEKR DPQEEDVDIF PSWTMSDEKV DRKSANIQKN ETVKVNRKSA 1020 GKRKMTYGRE TIKKAKLVES EDMVSDASVE DCIHQHHSIL RNKQSKFVEC NDPMSDDSVE 1080 DDSSRKHGVP VSKGAPYFGT DDTGSDDSLG DRHTLHRGFS GFKLPRWGEI EPSVSDDSLE 1140 HYSSQHRGKN IKSRTGKYIE RQDALSDECL ESGSLKQYRR IPKSKQTKVL KKNAILHDIR 1200 DDSFLWHHQK PSRIKKAKFI ETEDAVSEHS LENSSHQHRS MPQIKPAKHT AWEDAFSDDP 1260 DEDDNSLLQH RNVRSNMQFR EITSDDQLDD GANQYSRRVL RRKPVKTETI SQMKQEILRP 1320 VKRGASQTLK EEFAQSLKRG GRHTLKLETP QPKIHHATNR RGKRNEKLTD LESEDEQPGG 1380 PSTRLRKRTP KPTKLSEAKV KDKKPVAKKK MKTGSSLKTP AGHRDSKARD EESEYLCDIE 1440 GCNMSFGTKQ ELALHKRNIC PVKGCVKKFF SHKYLVQHRR VHMDDRPLKC PWKGCKMTFK 1500 WAWARTEHIR VHTGARPYVC AEPGCGQTFR FVSDFSRHKR KTGHSTKKGR G* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 1e-76 | 15 | 379 | 6 | 346 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 1e-76 | 15 | 379 | 6 | 346 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1385 | 1411 | RKRTPKPTKLSEAKVKDKKPVAKKKMK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
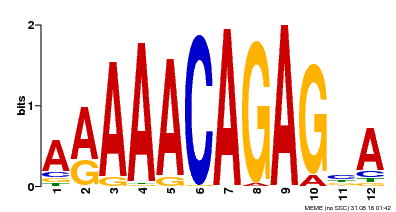
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681932 | 0.0 | LN681932.1 Cucumis melo genomic scaffold, anchoredscaffold00001. | |||
| GenBank | LN713266 | 0.0 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011651913.1 | 0.0 | PREDICTED: lysine-specific demethylase JMJ705 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A0A0LWI2 | 0.0 | A0A0A0LWI2_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004157814.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5648 | 32 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.391680.1 |



