 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.396070.1 | ||||||||
| Common Name | Csa_2G357210, LOC101223043 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 412aa MW: 44351.8 Da PI: 10.1423 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 48.1 | 2.6e-15 | 333 | 385 | 5 | 57 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkeva 57
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++eN++L+++ e+ ++ +
Cucsa.396070.1 333 RRQRRMIKNRESAARSRARKQAYTMELEAEVAKLKEENQELRQKQAEIMEMQK 385
79****************************************99999988865 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 1.1E-14 | 326 | 380 | No hit | No description |
| SMART | SM00338 | 1.6E-12 | 329 | 393 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.461 | 331 | 382 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.19E-10 | 333 | 381 | No hit | No description |
| CDD | cd14707 | 1.38E-25 | 333 | 385 | No hit | No description |
| Pfam | PF00170 | 3.4E-13 | 333 | 385 | IPR004827 | Basic-leucine zipper domain |
| PROSITE pattern | PS00036 | 0 | 336 | 351 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 412 aa Download sequence Send to blast |
MNFKDFGNDP SGGNGGGGGR PPANYPLARQ SSIYSMTFDE FQSMGSIGKD FGSMNMDELL 60 KNIWSAEEMQ TMASSAAAVG KEGGGSAGGS SGYLQRQGSL TLPRTLSQKK VDEVWKDIIN 120 EHASAKDGAT LAPNLQQRQQ TLGEVTLEEF LFRAGVVRED TQVTANPNNG GFFGNNTGFG 180 IAFQRQPKVP ENNNHIQIQS SNLSLNANGV RAHQPQPIFP KQATLTYGSQ LTLPSDAQLA 240 SPGIRGGIMG IADQGLNTNL MQGTALQGGR MGVVNIAAAP LPIATESPGN QLSSDGIGKS 300 NGDTSSVSPV PYALNGGIRG RRGNGIVDKV VERRQRRMIK NRESAARSRA RKQAYTMELE 360 AEVAKLKEEN QELRQKQAEI MEMQKNRALE VMDKQQGIKK RCLRRTQTGP W* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
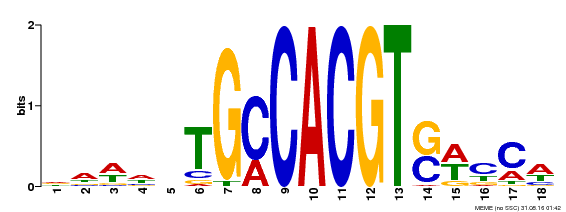
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681823 | 0.0 | LN681823.1 Cucumis melo genomic scaffold, anchoredscaffold00014. | |||
| GenBank | LN713257 | 0.0 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142865.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Refseq | XP_011649580.1 | 0.0 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Swissprot | Q9M7Q4 | 1e-131 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | A0A0A0LL86 | 0.0 | A0A0A0LL86_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004142865.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1962 | 34 | 81 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 1e-104 | abscisic acid responsive elements-binding factor 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.396070.1 |
| Entrez Gene | 101223043 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



