 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_000385 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 300aa MW: 33027.4 Da PI: 9.7518 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.8 | 3.9e-16 | 17 | 64 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W +Ed ll ++++++G g W++++r+ g++R++k+c++rw++yl
DCAR_000385 17 KGAWGSDEDALLRKCIEKYGEGKWHLVPRRAGLNRCRKSCRLRWLNYL 64
799*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 39.1 | 1.8e-12 | 70 | 113 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg + ++E +l +++k+lG++ W++Ia +++ gRt++++k++w++
DCAR_000385 70 RGDFAADEVDLMMRLHKLLGNR-WSLIAGRLP-GRTANDVKNFWNT 113
677889999*************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 21.921 | 11 | 68 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-20 | 13 | 66 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.95E-26 | 15 | 111 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.3E-13 | 16 | 66 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 17 | 64 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.04E-10 | 19 | 64 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-20 | 67 | 119 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.326 | 69 | 119 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.1E-10 | 69 | 117 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.5E-11 | 70 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.61E-7 | 75 | 115 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 300 aa Download sequence Send to blast |
MHPKALKSSS NPLKLRKGAW GSDEDALLRK CIEKYGEGKW HLVPRRAGLN RCRKSCRLRW 60 LNYLRPTIKR GDFAADEVDL MMRLHKLLGN RWSLIAGRLP GRTANDVKNF WNTNVQKKLT 120 TSSNHGQTEV VKAQEVVNKN QTSNAGTSAA TTHVVVKPLP RTLSKGTSVP CYNPNAIGHK 180 LSPWPGGMVY NKISSSNNNN NSCMAMNKTL SPAAALPDQD GTEWWKNLFA EIGIQGQEEG 240 SLEGHLVASS SGSENLEAER GLIRKMDESF APATEAGLVG DDGRSCWSDI WDLLNLDYS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-22 | 15 | 121 | 5 | 110 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH002/EGL3/MYC146, BHLH012/MYC1, or BHLH042/TT8. {ECO:0000269|PubMed:15361138}. | |||||
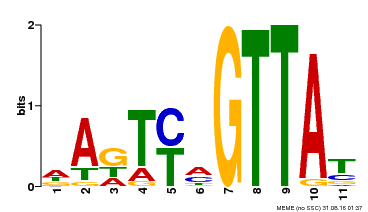
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00213 | DAP | Transfer from AT1G66370 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017228240.1 | 0.0 | PREDICTED: transcription factor MYB113-like | ||||
| Swissprot | Q9FNV9 | 2e-62 | MY113_ARATH; Transcription factor MYB113 | ||||
| TrEMBL | A0A175YA44 | 0.0 | A0A175YA44_DAUCS; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12 | 24 | 2154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G66370.1 | 8e-64 | myb domain protein 113 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_000385 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



