 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_002547 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 976aa MW: 107974 Da PI: 6.4106 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.2 | 3.2e-40 | 126 | 203 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C+adls+ak+yhrrhkvC vhska++++v +qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
DCAR_002547 126 VCQVDDCRADLSSAKDYHRRHKVCAVHSKATKAMVGSAMQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTHP 203
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.0E-32 | 120 | 188 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.735 | 124 | 201 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 7.98E-37 | 126 | 204 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.0E-29 | 127 | 200 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 3.50E-7 | 742 | 861 | No hit | No description |
| SuperFamily | SSF48403 | 4.82E-9 | 744 | 863 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.3E-6 | 755 | 865 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.549 | 801 | 822 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 976 aa Download sequence Send to blast |
MEWDLNDWKW DGDQFTAAPL RSVAADCRSR QFFPIGPEIP VVTGLSNSSS SCSEEIDLRN 60 EREQRELEKR RRVFVVDDDE LDDESGSLNL NLGGMVYPVA NGEVENWNGK SGKKTKLGGS 120 ASSGAVCQVD DCRADLSSAK DYHRRHKVCA VHSKATKAMV GSAMQRFCQQ CSRFHLLQEF 180 DEGKRSCRRR LAGHNKRRRK THPENSNTGV PVNDELSNSY LLISLLRILS NMHTDSSDQM 240 KDQDLLSHLL KNLASVAGTI NEGNLSGLLN GSSKLQLQNF VASRGQQLSR PTGQCTVMPS 300 SGMTQKREFV DNVSGGKFQT PPAQLSNIHF PTKDCIAANA NASNAKMEST KELNFDLNDV 360 YDDSEECMEP LERSDAPICV ENGSAGYPIW IHQDSDKSSP PQTSGNSGSL STQSPSSSSG 420 EAQLVQVRTD RIVFKLFGKN PSEFPLVLRS QILGWLAHSP TDIESYIRPG CVILTIYLRM 480 EKSTWEVVCS DLSFSLRKLL DSDSFWKEGW IYTRVQNRVA FVHDGEVVLD TQLTVTNDKN 540 CNISSIRPIA VTVSENANFL VKGSNMSQST RILCALEGMY LVQQSCSELM DGCGSLSESD 600 KGQSLSFPCS IPNVMGRGFI EVEDQTLGTS FFPFIVAEKD VCSEICTLES ALELAETTSG 660 IKGETEHLEV HDQALEFVHE MGWLLHRTQL KIRLGPNDPN LDLFSFKRFR YLMEFSLDHD 720 WCAVVKKLLC ILFSGIVDSG EHPNIERALL DIGLLHRAVQ RNCRPMVQAL LSFIPANVID 780 KYGPEQTLPG HIFRPDSAGP GGLTPLHIAA SSGGYETVLD ALTDDPQMVG LEAWKNARDG 840 AGLTPHDYAF QRGCHSYINM IQKKTKTRSG QGHVVVDIPG TIKQKLPGGP KPTEVSSFQT 900 EKAIMKLVQS NCNLCEQKLA YGNYRRSLAS YRPAMLAMVA IAAVCVCVAL LFKSSPEVLY 960 VFQPFRWEHL KFGSS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-31 | 118 | 200 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 68 | 72 | KRRRV |
| 2 | 195 | 201 | KRRRKTH |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
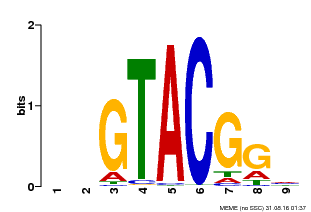
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017226642.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A169WU80 | 0.0 | A0A169WU80_DAUCS; Uncharacterized protein | ||||
| STRING | VIT_05s0020g00700.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1871 | 24 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_002547 |



