 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_010064 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 486aa MW: 53688.3 Da PI: 5.9699 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 50.1 | 6.4e-16 | 43 | 87 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT eE+ ++++a k++G W++I +++g ++t+ q++s+ qk+
DCAR_010064 43 RERWTDEEHHKFLEALKLYGRA-WRRIEEHIG-TKTAVQIRSHAQKF 87
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.56E-16 | 37 | 91 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.841 | 38 | 92 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.4E-9 | 39 | 94 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.4E-16 | 41 | 90 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.6E-11 | 42 | 90 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.8E-13 | 43 | 86 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.11E-9 | 45 | 88 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 486 aa Download sequence Send to blast |
MTSPDTLTAG LQSPADTQDK CQVVSSDDYA IKVRKQYTIT KQRERWTDEE HHKFLEALKL 60 YGRAWRRIEE HIGTKTAVQI RSHAQKFFSK VVRESSINEK NTVKPIEIPP PRPKKKPMHP 120 YPRKMVGRVE PGAHLSDELG RSASPSHSVS GRENRSPASV LSAVGSEATC VTDSNMQNCI 180 FSASSSDADD QTNSVFPSDT TPLPEENRSE NGSNSPIQAN DSSNEDEHVP TKLELFEADG 240 SFVKEESNDE VSTHSVKLFG KTVTVAESSL SSSRNNSCGS SHLESADERP VQTSPWNISQ 300 GMSYASPDAA EYTWTGVPCK APAVYYMQYA INNSINVDDE SSVPVPCWTF YGGVPYPFLQ 360 LRNSVQERVY MHFDGSNVQD KVLQKPGSLT GSNSGSINVE TDGDKTWEME TQCSQSLSEK 420 KDAEQKLTSP LKVRADAGFT EQKSILAKAN HGKGFVPYKR CLAERDNGTS ISGEEREERR 480 TRLCL* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
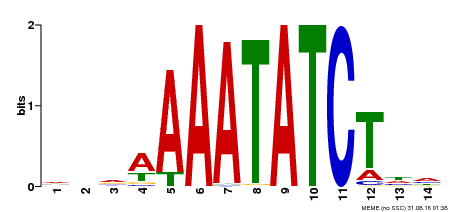
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017237482.1 | 0.0 | PREDICTED: protein REVEILLE 1-like | ||||
| TrEMBL | A0A166AIN5 | 0.0 | A0A166AIN5_DAUCS; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3185 | 24 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 3e-46 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_010064 |



