 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_010178 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 570aa MW: 62971.6 Da PI: 7.3581 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 55.4 | 1.1e-17 | 267 | 316 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr l+P++ ++K +Ka+ L ++veYI++Lq
DCAR_010178 267 RSKHSATEQRRRSKINDRFQMLRGLIPHS----DQKRDKASFLLEVVEYIQYLQ 316
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 1.4E-23 | 262 | 333 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.62E-19 | 263 | 336 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.348 | 265 | 315 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.26E-15 | 267 | 320 | No hit | No description |
| Pfam | PF00010 | 3.3E-15 | 267 | 316 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.2E-13 | 271 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 570 aa Download sequence Send to blast |
MELPQPRPYG SAEGRKSTHD FLSLYSPGQQ ESRSSQGGFL KTHDFLQPLE RVETDQGISK 60 EGNKGEPPME NSRPPALPAT NSVEHILPGG IGTYSISHIS YLNQRVPKPE GVDTPPAQAS 120 SSNRNDDNSN CSSYTGSGFT LWDESVVMNK GQTGKENNFA AKERHAVREV GVHVGGQWAT 180 SLERPSQSSS SHSFLNKAFS SLSSSQQSFT KKQSFMDMIK SAKNGQEDEE EEDDEFVIKK 240 ELSPHNKGNI SIKVDGKGTD QKPNTPRSKH SATEQRRRSK INDRFQMLRG LIPHSDQKRD 300 KASFLLEVVE YIQYLQEKVQ KYEDPYHRWS QEPPKMMPWN NSHRTVETFD DQSGGPNCGS 360 GPPLMFATKF DEANTNISST MPRGGQGLSE YEISTADTVR EMGHHRVLAN GGMTLHVPPQ 420 QTMYSHKGSS SAAVTLPPRV ASNTEIRTSH PHPQLWQSKP STTESTTVRE KIKDQEMTIE 480 SGTISISSVY SKELLNSLTQ SLQSSGVDLS QASISVQIDI ANQAKRNLNP STPVVQSDDP 540 PNLTMLQSRL PSGRGESEQA FKRLKTGRS* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
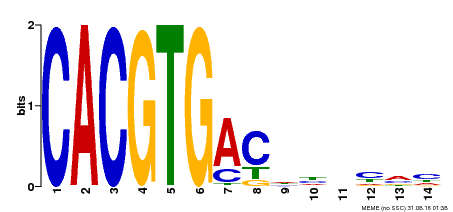
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017243146.1 | 0.0 | PREDICTED: transcription factor BIM1 | ||||
| TrEMBL | A0A166AKX6 | 0.0 | A0A166AKX6_DAUCS; Uncharacterized protein | ||||
| STRING | VIT_17s0000g04790.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3969 | 23 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.5 | 9e-87 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_010178 |



