 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_021667 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 370aa MW: 41724.7 Da PI: 9.488 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 48.5 | 1.5e-15 | 145 | 237 | 5 | 98 |
-..-HHHHT..T-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 5 ltpsdvlks..grlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
++ s++ ++++pk+f e++ ++++ +l+l +grsW v ++++ k +++ +++GW F+ +n L++gD++vF+l+++s+++++v + r
DCAR_021667 145 MKESHMVGGgwPNVYIPKSFKEAYTWQSNQ--KLILV-VEGRSWVVFCNMNSKCNQCRISRGWTIFAGDNSLRVGDVCVFELISSSSKKFKVFISR 237
555555555333479*********555333..45544.489******************************************9988877777766 PP
| |||||||
| 2 | B3 | 61.2 | 1.8e-19 | 275 | 366 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE- CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeehggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfr 98
ff+v+++ ++l g +++p +f+e++ k s ++l+ ++gr+W+vk+ ++kk +++GWk+F+ +n+L +gDf+vF+l++ e+ l+v +fr
DCAR_021667 275 FFSVEVHRSYLYGGSMTVPLDFIESNITK--DSCRVVLQLPDGRVWSVKCYINKKCA--KFSAGWKNFATENNLAAGDFCVFELVK--ERLLNVVIFR 366
88999999999**************9544..4557***************4455554..4899********************987..4446888887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 3.1E-4 | 33 | 62 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.5E-19 | 133 | 239 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 4.71E-18 | 134 | 238 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.27E-17 | 140 | 237 | No hit | No description |
| Pfam | PF02362 | 6.0E-14 | 141 | 237 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.7E-5 | 141 | 239 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.998 | 158 | 239 | IPR003340 | B3 DNA binding domain |
| Gene3D | G3DSA:2.40.330.10 | 1.3E-24 | 268 | 366 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 2.16E-25 | 269 | 367 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.81E-19 | 274 | 366 | No hit | No description |
| Pfam | PF02362 | 4.9E-18 | 275 | 366 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.917 | 275 | 368 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.3E-18 | 275 | 368 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0034613 | Biological Process | cellular protein localization | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 370 aa Download sequence Send to blast |
MTSSAISQGV FKDFRTEPQG QYNVEYFYSI SFGYLLVFEY KGDSKFQVLI FDPSASEIDY 60 SLARKDKPGS VKSAQVKVRQ IDIDDSSSSD DLIRPCKKTK AEPSSEASPK LGQEKAKEAS 120 KVDKDRALAV ANAFKSKNVS FIHVMKESHM VGGGWPNVYI PKSFKEAYTW QSNQKLILVV 180 EGRSWVVFCN MNSKCNQCRI SRGWTIFAGD NSLRVGDVCV FELISSSSKK FKVFISRASK 240 ETNCEENERL PRVRSEADKA RVLQSLKAYK PNRPFFSVEV HRSYLYGGSM TVPLDFIESN 300 ITKDSCRVVL QLPDGRVWSV KCYINKKCAK FSAGWKNFAT ENNLAAGDFC VFELVKERLL 360 NVVIFRVES* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4i1k_A | 8e-19 | 252 | 367 | 25 | 142 | B3 domain-containing transcription factor VRN1 |
| 4i1k_B | 8e-19 | 252 | 367 | 25 | 142 | B3 domain-containing transcription factor VRN1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00192 | DAP | Transfer from AT1G49480 | Download |
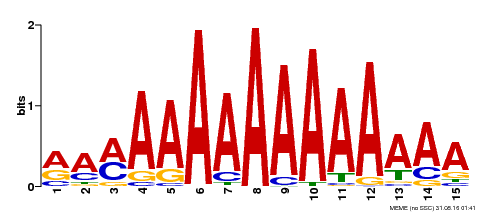
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017256890.1 | 0.0 | PREDICTED: B3 domain-containing transcription factor VRN1-like | ||||
| TrEMBL | A0A164VWN7 | 0.0 | A0A164VWN7_DAUCS; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA759 | 18 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49480.1 | 7e-20 | related to vernalization1 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_021667 |



