 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_023944 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 451aa MW: 49733.5 Da PI: 6.656 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 28.4 | 3.5e-09 | 262 | 313 | 12 | 63 |
HHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 12 kNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
NR +A rs +RK +i+eLe+kv++L++e ++L+ +l +l+ l++e+
DCAR_023944 262 ANRQSAARSKERKMRYIAELERKVQALQTEATTLAAQLTMLQRDTNGLTAEN 313
5****************************************99888887776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 5.6E-10 | 252 | 315 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.852 | 256 | 316 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.27E-9 | 257 | 306 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 6.9E-7 | 258 | 303 | No hit | No description |
| Pfam | PF00170 | 5.4E-7 | 259 | 309 | IPR004827 | Basic-leucine zipper domain |
| CDD | cd14703 | 3.75E-21 | 259 | 305 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 451 aa Download sequence Send to blast |
MEKDRSVNLG GGLLPPSGRF SPFRPSGSAL NVKTEPSGST AVPQLRPDSL AEAGRFGHGL 60 SSDTNHFSYD ISQMPDNPPK NRGHRRAHSE ILTLPDDLSF DSDLGVVGAL DGPSYSDETE 120 EDLFSMYLDM DKFNASATSS FQVGEQSSAS GQPSILAPAS GQAAPSTDNM VFASTDRPRI 180 RHQHSQSMDG STTIKQEMLM SGTEDPSSIE AKKSMSAAKL AELALVDPKR AKSFYITILH 240 PCLLTCNLRG IILAWQRNRI WANRQSAARS KERKMRYIAE LERKVQALQT EATTLAAQLT 300 MLQRDTNGLT AENSELKLRL QTTEQQVHLQ DALNEALKEE IQHLKLLTGQ NIANGGSMVN 360 FPPYGASQQY YPTNQAMQSM LTAQQFQQLQ LHSQKQQNQF QQQQLLQQFQ QQQHQYREQH 420 LSQGGDMKMK GATSSGNLGE SSQDISMSKE * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
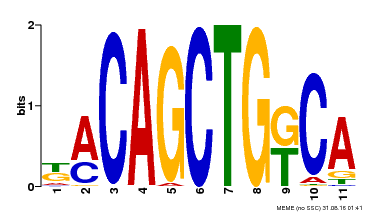
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017217544.1 | 0.0 | PREDICTED: probable transcription factor PosF21 | ||||
| Refseq | XP_017217545.1 | 0.0 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-146 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A161ZL68 | 0.0 | A0A161ZL68_DAUCS; Uncharacterized protein | ||||
| STRING | EMJ23579 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3965 | 21 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-132 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_023944 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



