 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_024178 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 818aa MW: 88592.1 Da PI: 6.6305 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 49.8 | 6.7e-16 | 488 | 525 | 2 | 39 |
TCR 2 ekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
+k+CnCkkskClk+YCeCfaag++C e C+C++C+Nk
DCAR_024178 488 ACKRCNCKKSKCLKLYCECFAAGVYCVEPCSCQECFNK 525
589**********************************8 PP
| |||||||
| 2 | TCR | 51 | 2.8e-16 | 573 | 611 | 1 | 39 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNk 39
++k+gCnCkks ClkkYCeC++ g+ Cs +C+Ce+CkN
DCAR_024178 573 RHKRGCNCKKSGCLKKYCECYQGGVGCSINCRCEGCKNA 611
589***********************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 9.7E-16 | 487 | 528 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 37.095 | 488 | 613 | IPR005172 | CRC domain |
| Pfam | PF03638 | 1.3E-11 | 490 | 525 | IPR005172 | CRC domain |
| SMART | SM01114 | 7.5E-18 | 573 | 614 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 8.0E-12 | 575 | 611 | IPR005172 | CRC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009934 | Biological Process | regulation of meristem structural organization | ||||
| GO:0048444 | Biological Process | floral organ morphogenesis | ||||
| GO:0051302 | Biological Process | regulation of cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 818 aa Download sequence Send to blast |
MGKDGVNLEK EGEGVVMDTP ERKQFTSPVS KIEDSPVFNY LNSLSPIKPF KSIHITQTFN 60 SLTFSSPPSV FTSPHVSSQR DSRFLRRHQF SDPSKPDFSG NSEEKVAKNE EATENAHNSA 120 QQQHFNQGSL ASESSAEPLY DTSVVVELPQ NLNGECVSPD CDGTSNCGMG IRCVSELGGS 180 STEVIPYAEI GSGKENIERE IHLEGPSHID ANKEAAGCDW DNLFSDEGDL LLFDTPNDSK 240 SLVDPGQRSL LPGMSFCTSL TDNMQISEAV NAVGLGPDNH SSLLECGQNI KAVIDQDIPA 300 SGSQSVAGDT SEKVDNEIAS NLQRGMRRRC LVFEMVGSRR KRLDDGSSTS VLLQCHENPA 360 SSDKCVVPLQ PVSNIPRRRL PGIGLHLNTL AATSVDHKVV KHEALVPGGQ PISVSGSAAY 420 INSSNSQQIL DSALVMYSSD RDMCPIGDGT PMAEDAAQGS GYMVNESLNH NSPKKKRRKS 480 DSDGESDACK RCNCKKSKCL KLYCECFAAG VYCVEPCSCQ ECFNKPIYED TVLATRKQIE 540 SRNPLAFAPK VIRNSDSIIE IGEETSKTPA SARHKRGCNC KKSGCLKKYC ECYQGGVGCS 600 INCRCEGCKN AFGRKDGTEV ELGEEEEAEH ENNAVNRSLQ ISTIQNDAEQ NPSFAIPATP 660 SQFGSQSIQP ISSKSKRPPR SSFLSIGGSS SAIYGTPAFG KLNPLRPLSK IQNDQKDEIP 720 EILQKNASPN SRVKLSSPNS KRVSPPHGNI GQSQGQRSSL IAISVCSLSK MLAMSSKFCV 780 QLLCVYRTRS YYFNVDAAGA CHRFRNIDAT TNNARIK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 4e-17 | 490 | 610 | 12 | 120 | Protein lin-54 homolog |
| 5fd3_B | 4e-17 | 490 | 610 | 12 | 120 | Protein lin-54 homolog |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 326 | 341 | RRRCLVFEMVGSRRKR |
| 2 | 473 | 477 | KKKRR |
| 3 | 473 | 478 | KKKRRK |
| 4 | 474 | 478 | KKRRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00370 | DAP | Transfer from AT3G22780 | Download |
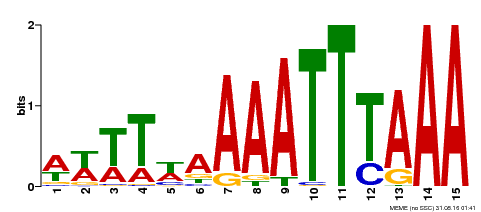
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017217612.1 | 0.0 | PREDICTED: protein tesmin/TSO1-like CXC 3 | ||||
| TrEMBL | A0A164T4E4 | 0.0 | A0A164T4E4_DAUCS; Uncharacterized protein | ||||
| STRING | XP_008224266.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA2467 | 23 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22780.1 | 1e-121 | Tesmin/TSO1-like CXC domain-containing protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_024178 |



