 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_026357 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1086aa MW: 119851 Da PI: 7.7383 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 126.6 | 9.8e-40 | 167 | 243 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
Cqv++C++dls+ak+yhrrhkvCevhska++++v +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+ rrk+q+
DCAR_026357 167 CQVDDCKEDLSTAKDYHRRHKVCEVHSKATKAIVGKQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRGRRKTQP 243
**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.0E-34 | 161 | 228 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.205 | 164 | 241 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.57E-36 | 165 | 245 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.7E-29 | 167 | 240 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1086 aa Download sequence Send to blast |
MEEVGTQVAS SHYIHQSLSK HFLDSQHHPM AAAKKRSMPF HPLGFHLPQS QQQQPMFGSG 60 FQQSRGQWHP NGWDWDSAKF LAKPVESDVI RGGPSTMSVQ SVMQRSHGDE IIAGNSVDLR 120 GNHVVEDDEN LLLELRGSRV NFSEENVLRP NKRVRSGSPA GGNYPKCQVD DCKEDLSTAK 180 DYHRRHKVCE VHSKATKAIV GKQMQRFCQQ CSRFHPLSEF DEGKRSCRRR LAGHNRGRRK 240 TQPDDVTSKF LPPTNRENVS NGDVDLVNLL AILARAQGNT EDSSINGSSI PNKDQFIQIL 300 QKINSLPLPA DFAAKLPPLG VTSNIAPFQS ESENKLNGNN SPSSTLDLLA VLSGNQAGSS 360 PDATAVPSQR SSHGSDSEKT RSPCTNMQTR HQNEFVSVGE RSSTSYQSPT ECSDGQVQEI 420 RTNLPLQLFS SSPESDSPPK LASSTKYFSS GSSNPMEERS PSSSPPFVQK LFPTVSSREA 480 VNPKRLLNRL EFNGSSKAGK DKGCSTSLDL FGGTNKCVDD SSVQSLPYQA GYTSSSGSDH 540 SPSSLNSDAQ NRSGRIYFKL IDRDPSQLPG KLRTQIFNWL GQSPSEMESY IRPGCVVLSI 600 YISMPSSAWE QLEENLLQNI TSLVQDSEDP FWRKGRFLVN TGRQLASYQD GKAHLRKATR 660 AWSFPEVVSV SPLAVVGGQE TTILLKGRNL SDQYTKVFCT HAVGYKLEET SGSASDDTTY 720 DSITLRNFTV ADEAPGILGR CFIEVENGFR GTSFPIIIAN ATICKELNLL ESAFDKVAAP 780 RDAISEESFL DSGRPISRED VLHFLNELGW VLQRKRYMSM FEVPEYKLHR FKFLFIFSVE 840 HDFCALVKTL LDVLLEICLG RDELSRESLQ MLLEIHLLNR AVKMRSRKMV DLLINYFVPT 900 DSGKTYIFLP NLAGPGGITP LHLAACMSDA NSMVDTLTTD PLEIGLHGWD SLLDANGLSP 960 CAYAQMRNNH SYNALVARKL VDRKNGQVSV PVGDEIQEQS LIAGQIHQAS FQIRQGQRSC 1020 SKCAVGAARY NRKTSASQGL LHRPYIHSML AIAAVCVCVC LFLRGAPDIG LVAPFKWENL 1080 GYGAV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 9e-29 | 157 | 240 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
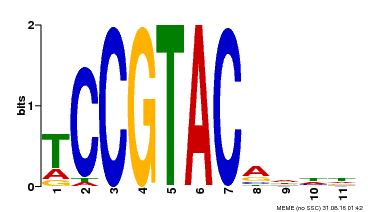
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017218515.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A161ZP29 | 0.0 | A0A161ZP29_DAUCS; Uncharacterized protein | ||||
| STRING | VIT_18s0122g00380.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12887 | 12 | 19 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_026357 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



