 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_026483 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 391aa MW: 44627.9 Da PI: 6.2978 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 34.1 | 6.2e-11 | 341 | 389 | 3 | 48 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqkyl 48
W+ E++ l +v+++G g+Wk+I ++ Rt+ ++k++w++++
DCAR_026483 341 LWSNVEEDTLRTGVRKYGVGNWKLILNMYRdifDDRTEVDLKDKWRNLM 389
5**********************************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.113 | 333 | 390 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-12 | 336 | 389 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 8.22E-15 | 336 | 389 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.0E-6 | 338 | 390 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-9 | 341 | 388 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 7.75E-16 | 342 | 389 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 391 aa Download sequence Send to blast |
MDPDIARWVL DFLVRQPLDD STLTSLLTTL PLSNTDTHLT KLLLLKRIDS EISKNAVSES 60 VLGLLEQMEE LDYREHVEIS DSMKRAYCIV AVDCTVRFLE EGEKGFYFEA VKRIWRTRIG 120 CMMNFERVGL VSEELERWKD EIESAVWDDG ACKVLLEKWG GLDVVESVRV YIEEARGRIG 180 PSFLELVTES LSGEAVKEVF GIDKKDIHKE KVQRKSKPSA VKQHKKFSSK TTRGVKIVDN 240 DDLTLEQQES HNKDSSLSPT DVAKFQEGLR TSTSSLGKKA EDPFPDTVNI SKNVASNMDS 300 VDGCTETGNL GRLHLPSPKE RVVSPLKKYE SPKPLRRRRR LWSNVEEDTL RTGVRKYGVG 360 NWKLILNMYR DIFDDRTEVD LKDKWRNLMA * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 334 | 340 | LRRRRRL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
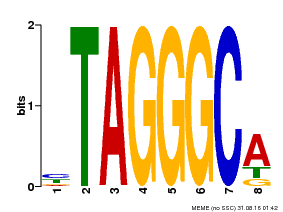
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017220376.1 | 0.0 | PREDICTED: uncharacterized protein LOC108197305 | ||||
| TrEMBL | A0A175YR22 | 0.0 | A0A175YR22_DAUCS; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3774 | 24 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 4e-27 | TRF-like 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_026483 |



