 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | DCAR_027015 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; campanulids; Apiales; Apiineae; Apiaceae; Apioideae; Scandiceae; Daucinae; Daucus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 644aa MW: 73143.1 Da PI: 5.9033 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 164.7 | 1.6e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcy 99
+e k+rwl+++ei+aiL n++++++ + + p+ g ++L++rk++r+frkDG++wkkkkdgktv+E+he+LKvg+ e +++yYah+e+nptf rrcy
DCAR_027015 30 EEaKSRWLRPNEIHAILCNHKHFTIYVRPVNLPPAGMMVLFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGNDERIHVYYAHGEDNPTFVRRCY 127
4559********************************************************************************************** PP
CG-1 100 wlLeeelekivlvhylevk 118
wlL+++le+ivlvhy++++
DCAR_027015 128 WLLDKKLEHIVLVHYRDTH 146
****************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.08 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 8.8E-73 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.2E-46 | 31 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.27E-11 | 381 | 466 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 9.83E-9 | 565 | 636 | No hit | No description |
| SuperFamily | SSF48403 | 3.11E-9 | 568 | 636 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 7.5E-8 | 576 | 637 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 10.949 | 583 | 636 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 644 aa Download sequence Send to blast |
MEGAVPVRLV GSEIHGFHTM ADLDIGNVME EAKSRWLRPN EIHAILCNHK HFTIYVRPVN 60 LPPAGMMVLF DRKMLRNFRK DGHNWKKKKD GKTVKEAHEH LKVGNDERIH VYYAHGEDNP 120 TFVRRCYWLL DKKLEHIVLV HYRDTHESQG SPAAPIPSNS CSILSEPPAS RILSEESNSV 180 VNEIYYTSEK AYLEPTETIQ DHETRLHEIN TLDWEELVVP EDPNKLNAPE TGILSHFEQQ 240 NPYEMNFHRN NSNLLSTNIY SAEHNSLEKF SDLVVGSNLD HSNTLSNVYS DIVDGQINSD 300 LQKNGHKALN AVCGDSLDTI SRNGFQTQDS FGRWMNCVIT DSPDSAFTGD RNLESSISNG 360 QETSALGDQQ QSSFSEQIFS ITDVSPASAL TTEETKILVV GYFHQAQSPF EKCNLFCVCG 420 DECVTAELVQ PGVYRSFVSP HEPGVFDLYL SFDGQNPISQ VVTFEYKSPP TEKLVNQSDD 480 ESKWKEFRYQ MRLAHLLFST SRRSNTLSTK LSPNALKEAK FFAQKASQIG SNWTSFLKCI 540 KENKLSFPQA KDNLFELTLQ NRLQEWLLER VVERCKATDY DDQGQGVIHL CAILGYKWAV 600 PQFVLSGLSL DYRDKFGWTA LHWAAYCGRL VVISFLSQLL FFF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
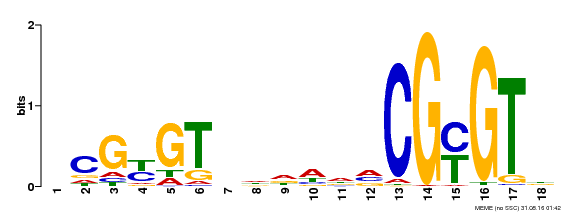
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017222907.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 6-like | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A175YQ31 | 0.0 | A0A175YQ31_DAUCS; Uncharacterized protein | ||||
| STRING | VIT_05s0077g01240.t01 | 0.0 | (Vitis vinifera) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4565 | 22 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | DCAR_027015 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



