 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca12447.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 794aa MW: 89172.7 Da PI: 6.9967 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 72.1 | 6.9e-23 | 101 | 202 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..E CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelv 93
f+k+lt sd++++g +++ +++a+e+ + + + +++l+ +d++ ++W++++i+r++++r++l++GW+ Fv++++L +gD ++F + +++el+
Dca12447.1 101 FCKTLTASDTSTHGGFSVLRRHADEClppldmSRQ-PPTQELVAKDLHRNEWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RGENGELR 196
99*****************************7333.34459************************************************..449999** PP
EEEE-S CS
B3 94 vkvfrk 99
v+v+r+
Dca12447.1 197 VGVRRA 202
***996 PP
| |||||||
| 2 | Auxin_resp | 119.4 | 2.9e-39 | 227 | 309 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha++t+++F+v+Y+Pr+s+++F+v+++++ +++k+++s+GmRfkm+fe+e+++e+r++Gt+vgv+d+d rW++SkWr+L+
Dca12447.1 227 AWHAIKTGTMFTVYYKPRTSPAQFIVPYDQYTESVKHHYSIGMRFKMRFEGEEAPEQRFTGTIVGVEDADANRWRESKWRCLQ 309
89********************************************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 9.9E-40 | 97 | 216 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 6.77E-40 | 97 | 230 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 3.93E-21 | 99 | 201 | No hit | No description |
| PROSITE profile | PS50863 | 11.251 | 101 | 203 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 9.8E-21 | 101 | 202 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.1E-22 | 101 | 203 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 9.7E-37 | 227 | 309 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 3.9E-10 | 655 | 758 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 23.537 | 668 | 752 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 1.37E-11 | 670 | 745 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 794 aa Download sequence Send to blast |
MSSTEVSTKQ NSEQFPPQIE TKVDPETALY RELWHACAGP LVTVPRINDL VYYFPQGHIE 60 QAEADTDEVY AQVMLQPQKE ENSVEKEPPA QPQPRFHVHS FCKTLTASDT STHGGFSVLR 120 RHADECLPPL DMSRQPPTQE LVAKDLHRNE WRFRHIFRGQ PRRHLLQSGW SVFVSSKRLV 180 AGDAFIFLRG ENGELRVGVR RAMRQQVIVP SSVISSHSMH LGVLATAWHA IKTGTMFTVY 240 YKPRTSPAQF IVPYDQYTES VKHHYSIGMR FKMRFEGEEA PEQRFTGTIV GVEDADANRW 300 RESKWRCLQV RWDETSTVPR PDRVSPWQVE PAVTPPTINP LPVSRVKRPR PNVATQSQDS 360 SAFTREGSSK LTADRSPASM LSRVLQGQEY PNMNPSLADS NESETVEKPI AWPNAVDEEK 420 TNSPSVSRKY VPEHWAPSMR PGPTYTDLLS GFGGHSDQPS GFHSSYQSAV ASPIKKSVVE 480 QDGRFNILGN PWSLNTVMSP GLSLNITDSS VKVPSQNSSI SYHVQGNGRY GSFGDIPVLH 540 GHRVDHHSGN WMMPPPGPPR FDNHPRSREM PMREPTKPKD GNCKLFGIPL ISSPVQQGSA 600 ILSRNGDESS NLLPPLLQHN GHHSEADRNS EQSKDCKQTE NLPTSNEQEK KLGSSRDCQA 660 KFHCGSTRSC TKVHKQGIAL GRSVDLSKFS DYDQFIDELD HLFDFNGELK APEKNWLIVY 720 TDDEGDMMLV GDDPWQEFCA MVRKIFIYTK EEVQKMTPKT LNSRTEDNPP AIGGSDSKEG 780 KQSLPSSASY AESH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-149 | 28 | 331 | 18 | 354 | Auxin response factor 1 |
| 4ldw_A | 1e-149 | 28 | 331 | 18 | 354 | Auxin response factor 1 |
| 4ldw_B | 1e-149 | 28 | 331 | 18 | 354 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 346 | 350 | KRPRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs) (By similarity). Could act as transcriptional activator or repressor (PubMed:26959229, PubMed:26716451, PubMed:27645097, PubMed:28167023). Involved in the control of fruit ripening process (PubMed:26959229, PubMed:26716451, PubMed:28167023). Regulates expression of a number of ripening regulators, transcription factors, and ethylene biosynthesis and signaling components (PubMed:26959229, PubMed:26716451). May act as a transcriptional repressor of auxin-responsive genes (PubMed:26716451). Regulates vegetative growth, lateral root formation and flower organ senescence, possibly partially by regulating gene expression of auxin and ethylene response factor (ERF) genes (PubMed:28167023). Plays a negative role in axillary shoot meristem formation (PubMed:27645097). {ECO:0000255|RuleBase:RU004561, ECO:0000269|PubMed:26716451, ECO:0000269|PubMed:26959229, ECO:0000269|PubMed:27645097, ECO:0000269|PubMed:28167023}. | |||||
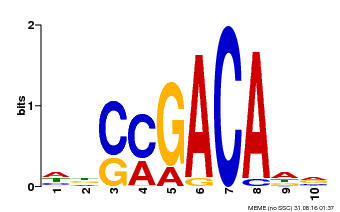
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By tomato fruit ripening. Expression in tomato fruits is up-regulated by exogenous application of ethylene and down-regulated by inhibition of ethylene receptors with 1-methylcyclopropene (1-MCP) (PubMed:26959229, PubMed:26716451). Up-regulated by plant hormone abscisic acid (ABA) in tomato fruits (PubMed:26959229). Expression is not up-regulated by plant hormone auxin in tomato fruits (PubMed:26716451). Up-regulated by auxin and gibberellic acid (GA), and down-regulated by ethylene in seedlings (PubMed:28167023). Down-regulated during decapitation-induced axillary shoot development and by excision of immature leaves. Down-regulation is inhibited by the application of auxin on the cut surface (PubMed:27645097). {ECO:0000269|PubMed:26716451, ECO:0000269|PubMed:26959229, ECO:0000269|PubMed:27645097, ECO:0000269|PubMed:28167023}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010678349.1 | 0.0 | PREDICTED: auxin response factor 2 | ||||
| Swissprot | Q2LAJ3 | 0.0 | ARF2A_SOLLC; Auxin response factor 2A | ||||
| TrEMBL | D7SH69 | 0.0 | D7SH69_VITVI; Auxin response factor | ||||
| STRING | XP_010678349.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



