 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca12805.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 299aa MW: 32244.8 Da PI: 6.6021 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 35.1 | 2.5e-11 | 130 | 175 | 5 | 55 |
HHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
h+++Er RR+ri +++ L+el+P+ + K +Ka++L + ++Y+k Lq
Dca12805.1 130 HSIAERIRRERIAERMKGLQELVPNgN------KTDKASMLDEIIDYVKFLQ 175
99***********************55......******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.31E-16 | 123 | 184 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.914 | 125 | 174 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.3E-15 | 127 | 184 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.2E-9 | 130 | 175 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.6E-13 | 131 | 180 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048767 | Biological Process | root hair elongation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 299 aa Download sequence Send to blast |
MEEELLQNQE VPPQHNNNNN NSNNNNNDDF LDQILSTLPN CPWLPPVLPS DEHHSAAAAS 60 PFHFAVNNNP LAGEDGSFQA MYNGFNGPFH DIGRPPNPPL TTPSTGGKSS GAAPNRQRVR 120 ARRGQATDPH SIAERIRRER IAERMKGLQE LVPNGNKTDK ASMLDEIIDY VKFLQLQVKL 180 LSMSRLGGAA ASAPLVADAP SENGGNGRNS NPNQSNDNIT TEHQVAKMME DDLGSAMQYL 240 QRKGLCLMPI SLATAISSAT CQPHNLSNTP SNTAPNMSGL TVQSAAAAME NTIKKASKH |
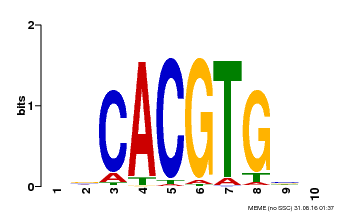
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold, UV, ethylene (ACC), flagellin, jasmonic acid (JA), and salicylic acid (SA) treatments. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023874690.1 | 1e-87 | transcription factor bHLH66-like | ||||
| Swissprot | Q9ZUG9 | 9e-82 | BH066_ARATH; Transcription factor bHLH66 | ||||
| TrEMBL | A0A2P5F3C2 | 9e-86 | A0A2P5F3C2_TREOI; Basic helix-loop-helix transcription factor | ||||
| STRING | XP_007138416.1 | 4e-85 | (Phaseolus vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G24260.1 | 2e-63 | LJRHL1-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



