 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca16138.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 327aa MW: 36201.7 Da PI: 6.7673 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 44 | 4e-14 | 43 | 92 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h+ E+rRR++iN++f+ Lr+l+P+ ++K + a+ L +++eY++ Lq
Dca16138.1 43 RSKHSVTEQRRRSKINERFQMLRDLMPHG----DQKRDTASFLLEVIEYVQLLQ 92
899*************************7....79****************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 2.0E-21 | 41 | 113 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 9.03E-18 | 41 | 112 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.535 | 41 | 91 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.15E-13 | 43 | 96 | No hit | No description |
| Pfam | PF00010 | 1.5E-11 | 43 | 92 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.2E-10 | 47 | 97 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MKSSKSHTTQ EDDDEYDDDF ASKKEGSSNI KDVKNNDKVS AIRSKHSVTE QRRRSKINER 60 FQMLRDLMPH GDQKRDTASF LLEVIEYVQL LQEKVQKYEG PYQQLGTEPS KLVPWRHSQW 120 RVQSYLGHPQ SVKNGFGSGS VSTLVGKTDD RSFSSNASLI SHLQTPSEMD SGHDNHRNLD 180 QHPDVGDKAA SVPTSFHMHA ASIGRSDGDD SCLSHRDASS IPRGIQSDDQ PGDHVIEGGT 240 ISISSAYSHG LLMTLTQALQ NSGVDLTEAS ISIRIDLGKR ASPNLLSSLS SNSKEHEFQV 300 SQDTKIRHAS DGNSRDGLDQ PHKKLKS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00219 | DAP | Transfer from AT1G69010 | Download |
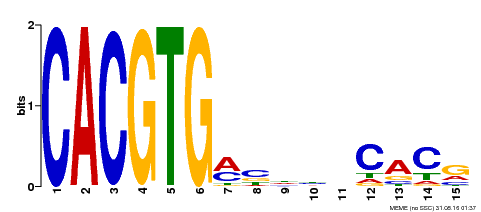
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by heat treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021756146.1 | 1e-135 | transcription factor BIM2-like | ||||
| Swissprot | Q9CAA4 | 1e-81 | BIM2_ARATH; Transcription factor BIM2 | ||||
| TrEMBL | A0A0K9RDJ7 | 1e-126 | A0A0K9RDJ7_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010681151.1 | 1e-132 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69010.1 | 2e-80 | BES1-interacting Myc-like protein 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



