 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca19176.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 864aa MW: 93670.5 Da PI: 6.072 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.2 | 9.3e-21 | 166 | 221 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Dca19176.1 166 KKRYHRHTPQQIQELESVFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 221
688999***********************************************999 PP
| |||||||
| 2 | START | 200.9 | 5.6e-63 | 376 | 601 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE.....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEE CS
START 1 elaeeaaqelvkkalaeepgWvkss.....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetlevi 85
ela++a++elvk+a+ +ep+W +s e+ n de+++ f + + + +ea+r++g+v+ ++ lve+l+d + +W+e+++ +a+t++vi
Dca19176.1 376 ELALSAMDELVKMAQTDEPVWLRSIdcggrEIFNHDEYMRCFNSCIGlkpngFVTEASRETGMVIINSLALVETLMDAN-RWTEMFPgmiaRASTTDVI 473
57899*************************************88777999999**************************.******************* PP
CTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX CS
START 86 ssg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgrlp 177
ssg galqlm aelq+lsplvp R + f+R+++q+ +g+w++vdvS+d ++++ + +v +++lpSg+++++++ng+skvtwveh++++++ +
Dca19176.1 474 SSGmggtrnGALQLMHAELQVLSPLVPvRAVNFLRFCKQHAEGVWAVVDVSIDCIRENSGAPTYVNCRRLPSGCVVQDMPNGYSKVTWVEHAEYDDSNI 572
**********************************************************99*************************************** PP
HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 178 hwllrslvksglaegaktwvatlqrqcek 206
h+l+r+l++ g+ +ga++w atlqrqce+
Dca19176.1 573 HQLYRPLISAGMGFGAQRWIATLQRQCEC 601
***************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.24E-20 | 153 | 223 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-21 | 162 | 231 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.362 | 163 | 223 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-17 | 164 | 227 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 8.60E-19 | 165 | 223 | No hit | No description |
| Pfam | PF00046 | 2.7E-18 | 166 | 221 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 198 | 221 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 44.598 | 367 | 604 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.09E-33 | 368 | 601 | No hit | No description |
| CDD | cd08875 | 9.37E-123 | 371 | 600 | No hit | No description |
| Pfam | PF01852 | 3.5E-55 | 376 | 601 | IPR002913 | START domain |
| SMART | SM00234 | 5.0E-46 | 376 | 601 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.74E-24 | 630 | 857 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 864 aa Download sequence Send to blast |
MSFGGFLNNC GGGSCGGSGG GNNVDSGGGG GGGRIGVGDI SYIHNNHSNN MPPGAITQSP 60 HLVGPTSSFN KSPSSFFNSP GLSLALQTGM EGQEVGRLRD HNNYEGNNNE SESNNNINNN 120 NNNNNGGVIG IRRSREEEHD SRSGSDNLEG ASGDDQDIAD LPPSRKKRYH RHTPQQIQEL 180 ESVFKECPHP DEKQRLELSK RLCLETRQVK FWFQNRRTQM KTQLERHENS ILRQENDKIR 240 AENMSIREAM RNPMCSNCGG PAIIGDISME EQHLRIENVR LKDELDRVCV LASKFLGRPI 300 SSLSTLSTMS GSIAPHMPNS ALELGVGMSN GYGNSMTTVS HTLPLAPDFC NGISPTKASG 360 SLGGIETSLE RSMYLELALS AMDELVKMAQ TDEPVWLRSI DCGGREIFNH DEYMRCFNSC 420 IGLKPNGFVT EASRETGMVI INSLALVETL MDANRWTEMF PGMIARASTT DVISSGMGGT 480 RNGALQLMHA ELQVLSPLVP VRAVNFLRFC KQHAEGVWAV VDVSIDCIRE NSGAPTYVNC 540 RRLPSGCVVQ DMPNGYSKVT WVEHAEYDDS NIHQLYRPLI SAGMGFGAQR WIATLQRQCE 600 CLAILMSSNV PSGDNTAITA SGRRSMLKLA QRMTNNFCVG VCASTVHQWN KLQMGNVDED 660 VRVMTRKSVD DPGEPPGIVL SAATSVWLPV SPQRLFDFLR DDRLRSEWDI LSNGGPMQEM 720 AHIAKGQDQG NCVSLLRAGA MNPNQSSMLI LQETCTDASG SLVVYAPVDI PAMHVVMNGG 780 DSAYVALLPS GFSIVPDGPA SSGSNSSGQI SGLNRVGGSL LTVAFQILVN NLPTAKLTVE 840 SVETVNNLIS CTVQKIKAAL HCEN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
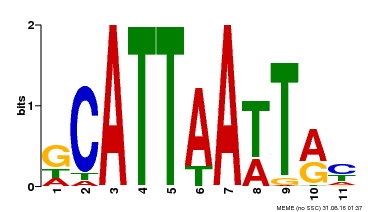
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010671720.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | A0A0K9QLI3 | 0.0 | A0A0K9QLI3_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010671720.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



