 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca20625.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 287aa MW: 33500 Da PI: 7.9852 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 20.9 | 9.2e-07 | 13 | 37 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp dC+ s++rk++L+rH+ H
Dca20625.1 13 FVCPvdDCNASYRRKDHLNRHLIQH 37
89*******************9877 PP
| |||||||
| 2 | zf-C2H2 | 15.2 | 6.1e-05 | 42 | 67 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
+kCp +C k F+ + n rH++ H
Dca20625.1 42 FKCPveNCEKEFTVQGNIARHLKEfH 67
89*********************988 PP
| |||||||
| 3 | zf-C2H2 | 18.8 | 4.5e-06 | 81 | 105 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ Cgk F+ s+L++H +H
Dca20625.1 81 HVCQknGCGKEFKYASQLQKHEESH 105
789999***************9988 PP
| |||||||
| 4 | zf-C2H2 | 12 | 0.00064 | 114 | 136 | 4 | 23 |
T..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 4 p..dCgksFsrksnLkrHirt.H 23
dC k Fs+++ L+ H+r+ H
Dca20625.1 114 AdpDCMKAFSNRECLQAHMRScH 136
455999***************66 PP
| |||||||
| 5 | zf-C2H2 | 17.7 | 9.8e-06 | 169 | 193 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C+ sF ++snL++H + H
Dca20625.1 169 KCNfdGCDHSFATSSNLRQHVKAvH 193
79999***************99877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF57667 | 8.29E-7 | 10 | 39 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.5E-10 | 10 | 39 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.5E-4 | 13 | 37 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 9.64 | 13 | 42 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 15 | 37 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.85E-6 | 40 | 68 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-5 | 40 | 64 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.032 | 42 | 72 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.026 | 42 | 67 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 44 | 67 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.7E-6 | 72 | 106 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.6E-6 | 75 | 105 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.554 | 81 | 110 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.004 | 81 | 105 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 83 | 105 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.91 | 111 | 136 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 8.829 | 111 | 136 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 113 | 136 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 17 | 139 | 160 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 1.63E-9 | 151 | 206 | No hit | No description |
| PROSITE profile | PS50157 | 12.05 | 168 | 198 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.6E-4 | 168 | 193 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 7.5E-8 | 169 | 193 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 170 | 193 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-4 | 194 | 222 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.8 | 199 | 225 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 201 | 225 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MVVLYSVFLQ RPFVCPVDDC NASYRRKDHL NRHLIQHEGK LFKCPVENCE KEFTVQGNIA 60 RHLKEFHEGK PYVRHEELKK HVCQKNGCGK EFKYASQLQK HEESHVESCE AFCADPDCMK 120 AFSNRECLQA HMRSCHQYII CEVCGSKHLR KNYKRHLRSH DDNTNEVLKC NFDGCDHSFA 180 TSSNLRQHVK AVHLNLRPFT CSFPGCGMSF AFKHVRDNHE KSFRHVHVPG DLEEFDEEWR 240 SRPHGGRKRK CPTVEMLMSK RVTPPNECDS VLNHSAEYLS WLLSADD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2i13_A | 4e-12 | 10 | 222 | 19 | 183 | Aart |
| 2i13_B | 4e-12 | 10 | 222 | 19 | 183 | Aart |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 241 | 249 | RPHGGRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
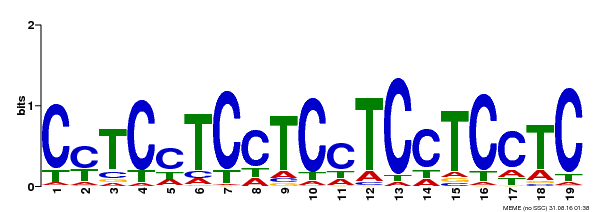
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021745786.1 | 1e-150 | transcription factor IIIA-like isoform X1 | ||||
| Refseq | XP_021745787.1 | 1e-151 | transcription factor IIIA-like isoform X2 | ||||
| Swissprot | Q84MZ4 | 1e-92 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0K9R4E8 | 1e-145 | A0A0K9R4E8_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010673198.1 | 1e-145 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.1 | 3e-95 | transcription factor IIIA | ||||



