 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca211.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 327aa MW: 36114.6 Da PI: 9.8313 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.2 | 9.1e-14 | 134 | 178 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + +q G+g+W+ I+r + k+R++ q+ s+ qky
Dca211.1 134 PWTEEEHRLFLVGLNQVGKGDWRGISRNFVKTRSPTQVASHAQKY 178
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50158 | 9.323 | 12 | 27 | IPR001878 | Zinc finger, CCHC-type |
| PROSITE profile | PS51294 | 17.246 | 127 | 183 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.22E-17 | 129 | 183 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.3E-15 | 130 | 181 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 5.6E-10 | 131 | 181 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.5E-10 | 133 | 178 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.2E-10 | 134 | 178 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.03E-9 | 134 | 179 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0031540 | Biological Process | regulation of anthocyanin biosynthetic process | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0080167 | Biological Process | response to karrikin | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MTGQSIKIPR TCSQCGNNGH NARTCPDHTV TPSNYNTNIN TNTSNGNNGV MLFGVRLTHS 60 VAENSTASPQ RPIESSSLSG FRKCMSMNNL AQYEQQQQQQ QQQQGNNCHD SGYASDDGIH 120 ASGGRNRERK RGVPWTEEEH RLFLVGLNQV GKGDWRGISR NFVKTRSPTQ VASHAQKYFL 180 RRTNLNRRRR RSSLFDITTD TMTRSASADQ LHNLDTTFQQ NQVTNQPTLF STPCTPTNLL 240 PNLAKHDNSM RTTAPVPPSV KMANLNLNQS PSSSPASSTS TNTTRERELE SFLSLKLEHA 300 PSDEAESSGL SNFGGFNGNR DNTISVA |
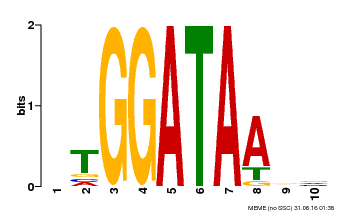
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00630 | PBM | Transfer from PK17526.1 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010683349.1 | 7e-90 | PREDICTED: transcription factor MYB1R1 | ||||
| TrEMBL | A0A0K9R9Z7 | 1e-86 | A0A0K9R9Z7_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010683349.1 | 3e-89 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G70000.2 | 5e-59 | MYB_related family protein | ||||



