 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca23162.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 327aa MW: 35276.4 Da PI: 9.4569 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 203 | 8.7e-63 | 2 | 149 | 1 | 149 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqsslk 99
+gsgr+ptw+ErEnnkrRERrRRa+aakiyaGLR++Gny+lpk++DnneVlkAL+r+AGw+vedDGttyrkg+kp e ++++g sas+sp+ss+
Dca23162.1 2 TGSGRMPTWRERENNKRRERRRRAVAAKIYAGLRMYGNYRLPKHCDNNEVLKALARDAGWIVEDDGTTYRKGCKPVEPMDIIGGSASVSPCSSYP---- 96
689********************************************************************************************.... PP
DUF822 100 ssalaspvesysaspksssfpspssldsislasa.....asllpvlsvlslvsss 149
+ +sy++sp+sssfpsp+++ +++ + +sl+p+l++ls++ ss
Dca23162.1 97 --VSPRAGASYNPSPASSSFPSPARSYYAPNLVNgttdpNSLIPWLKNLSSSGSS 149
..44556789*************9977665544446677*********9985544 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 6.7E-63 | 3 | 144 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MTGSGRMPTW RERENNKRRE RRRRAVAAKI YAGLRMYGNY RLPKHCDNNE VLKALARDAG 60 WIVEDDGTTY RKGCKPVEPM DIIGGSASVS PCSSYPVSPR AGASYNPSPA SSSFPSPARS 120 YYAPNLVNGT TDPNSLIPWL KNLSSSGSSS TSPKLPYIHI PGYSISAPVT PPLSSPTCRT 180 PRAKNDWDDN NVAPPWANPL NYSLPSSTPH SPGRKQQPPR GWLANIPNGP TSPTFSLVSS 240 NPFASKDEVM AGGGSRMWTP GQSGTSSPVV TTSFDRTADV PMSDGISAEF AFGNGMTGLV 300 KAWEGERIHE ECTPDDLELT LGSSKTR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 5e-23 | 6 | 87 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 5e-23 | 6 | 87 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 5e-23 | 6 | 87 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 5e-23 | 6 | 87 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
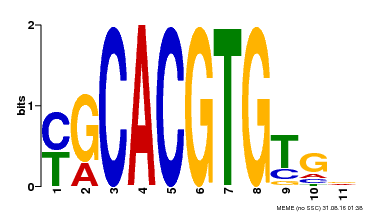
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010665681.1 | 1e-164 | PREDICTED: BES1/BZR1 homolog protein 4 | ||||
| Swissprot | Q9ZV88 | 1e-106 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A0J8B9H3 | 1e-163 | A0A0J8B9H3_BETVU; Uncharacterized protein | ||||
| STRING | XP_010665681.1 | 1e-164 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-87 | BES1/BZR1 homolog 4 | ||||



