 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca23634.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 482aa MW: 53574.6 Da PI: 6.7181 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 27.9 | 4.3e-09 | 306 | 353 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k Ka +Le+ ++Y++sLq
Dca23634.1 306 SHSLAERVRREKIGERMKLLQDLVPGC----NKVTGKALMLEEIINYVQSLQ 353
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 1.49E-9 | 300 | 357 | No hit | No description |
| SuperFamily | SSF47459 | 1.7E-16 | 300 | 370 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.396 | 302 | 352 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.4E-16 | 302 | 371 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.4E-6 | 306 | 353 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.9E-10 | 308 | 358 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
MDKNFFMDSS IAHQLQHDHP FSFDFFLDTQ RPHDHFDSEL SSMVSSPTSN SAISTDNFMI 60 SELIGKIGNI CNTSSLPNYI NSGNCSTNNS CYNTPLNSPP KIGKSTPNID HMVNENFSSL 120 GNLPAMPGGP GFAERAAKFS SFGSKSFNGR TGSSQFGSSN VNQNGNLGGE NGKLHRVCST 180 PLLDVEVTRS KIEVKKLENF VNDHQKVELS NNLENSSIST QIQVKENGSS RKRKATNVKG 240 KEKEKEIGCL NKVSENGDEA NAKRSKGGET TKLEENKGGE EDEKKNSEPF KDYIHVRARR 300 GQATDSHSLA ERVRREKIGE RMKLLQDLVP GCNKVTGKAL MLEEIINYVQ SLQRQVEFLS 360 MKLSNVNPRL ETSMSNSFLK DDNEPNCSME HPMYQTLYGQ PQQNNAQDTM QYSMDPFNPL 420 LNHNHSLHSS LIDGFSNYLS QLPSLCEGDL QSIVHMGFGQ KDNGFHAQTL PGTNQAQMKI 480 EF |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00660 | PBM | Transfer from Pp3c6_9520 | Download |
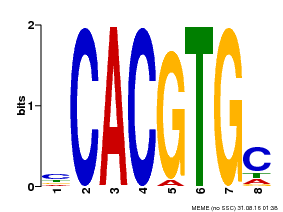
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010685111.1 | 1e-171 | PREDICTED: transcription factor bHLH78 | ||||
| TrEMBL | A0A0J8C085 | 1e-169 | A0A0J8C085_BETVU; Uncharacterized protein | ||||
| STRING | XP_010685111.1 | 1e-170 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G07340.1 | 1e-71 | bHLH family protein | ||||



