 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca28694.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 333aa MW: 36127.9 Da PI: 8.4826 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 25.4 | 3.3e-08 | 7 | 58 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT.......-H.HHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg.......tW.ktIartmgkgRtlkqcksrwqky 47
+WT eE++ + ++++++ + W ktI++ ++ +++ +q+++++q +
Dca28694.1 7 AWTREEEKAFENGIALHMSSldqiddeEWeKTISSLVP-TKSIEQIREHYQVL 58
6*****************99*******99679******.************86 PP
| |||||||
| 2 | Myb_DNA-binding | 40.3 | 7.4e-13 | 159 | 203 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ l++ + ++G+g+W++I+r + +Rt+ q+ s+ qky
Dca28694.1 159 PWTEEEHRLFLLGLDKFGKGDWRSISRNFVITRTPTQVASHAQKY 203
7*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 9.7E-4 | 4 | 61 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.7E-6 | 7 | 58 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.32E-7 | 7 | 59 | No hit | No description |
| PROSITE profile | PS50090 | 5.68 | 8 | 59 | IPR017877 | Myb-like domain |
| SuperFamily | SSF46689 | 6.14E-7 | 8 | 66 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.81 | 151 | 208 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.51E-17 | 154 | 209 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.1E-17 | 155 | 206 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.3E-10 | 156 | 206 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.5E-11 | 157 | 202 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 6.15E-10 | 159 | 204 | No hit | No description |
| Pfam | PF00249 | 1.9E-11 | 159 | 203 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MPNLAAAWTR EEEKAFENGI ALHMSSLDQI DDEEWEKTIS SLVPTKSIEQ IREHYQVLVD 60 DVNAIESGKV ALPNYGSNNN EEEVLLSNCS KDKNNNNSNS NNSSSHGQYS KEQKGNSNNN 120 NNNNNNNNNG NGFSGFGVGN NSASSKASKL DQERKKGIPW TEEEHRLFLL GLDKFGKGDW 180 RSISRNFVIT RTPTQVASHA QKYFIRLNSM NRDRRRSSIH DITSVNGGGL STNQVPITGQ 240 QNSAPTQAVG GAVTAGANKH RTHANVAGAP NLGMYGTQIG HPVSGAPVAP LIPGHMGSAV 300 GTPLMLPPPG HHPHHPHYAV PMAYPMAPPP MHR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00191 | DAP | Transfer from AT1G49010 | Download |
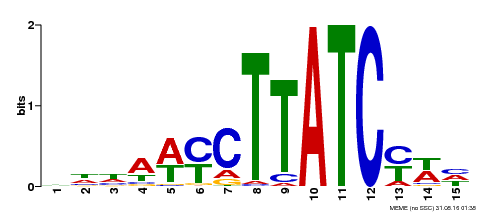
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021748976.1 | 1e-129 | transcription factor SRM1-like | ||||
| TrEMBL | A0A1S3UM88 | 1e-111 | A0A1S3UM88_VIGRR; transcription factor MYBS1 | ||||
| STRING | XP_010673224.1 | 1e-121 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08520.1 | 3e-43 | MYB family protein | ||||



