 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca37232.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 819aa MW: 93780.7 Da PI: 6.6543 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 82.3 | 8.3e-26 | 79 | 165 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
k+Y+eYAk++GF++++++s++sk+++ +++++f Cs++g + e++ + +r++++++t+Cka+++vk++k gkw+++++ +eHnHel p
Dca37232.1 79 KYYQEYAKSMGFTTSIKNSRRSKKSRDFIDAKFACSRYGVTPESDAA----SRRSSVKKTDCKASMHVKRNKCGKWVIHEFIKEHNHELLP 165
69***************************************999887....89999********************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.7E-23 | 79 | 165 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 2.0E-20 | 286 | 370 | IPR018289 | MULE transposase domain |
| Pfam | PF04434 | 5.5E-8 | 565 | 599 | IPR007527 | Zinc finger, SWIM-type |
| PROSITE profile | PS50966 | 10.324 | 565 | 601 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 2.6E-8 | 576 | 603 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 819 aa Download sequence Send to blast |
MEFLSTNGHP SVSNCVVDQG ENVHHGDEDD ENIGDLINEV HDQAVGTLSS TTNNLLLEGD 60 IEMGPDSDLQ FESHEAAYKY YQEYAKSMGF TTSIKNSRRS KKSRDFIDAK FACSRYGVTP 120 ESDAASRRSS VKKTDCKASM HVKRNKCGKW VIHEFIKEHN HELLPALAYY FRIHGNVKLA 180 EKSNIDILHA VSERTKKVYV DMSRQAAAVP NLESSRDILG SQLGKGNRLV LEEGDAQFLI 240 DYCMCVKKEN PKFFYAIDLD DKQYLRNFFW VDAKSRSDYI SFSDAISLDT SYIRTNEKLP 300 LIIFVGVNHH LQAMLLGCAL VSDDLTSNFV WLMKTWLRAM GGNAPSVILT DQHPSLKPAI 360 QEVFPDSRHC YHLCNVLEEI PKNLPHIIKH HGNFAAKFNK CVFKSWTDEQ FELRWWKMVG 420 RFDLQENEWF RSLYEDRKMW IPLYIDDAFL AGMSSVHQAE SVNSFFDKYI QRKLTLKEFV 480 KQYGLILQNR YEEEAIADFD TWHKQPALKS PSPWEKQMST LYTHAIFKKF QVEVLGVVGC 540 HPKKELEDGN SVTYRVEDCE KDETFFVTWN EGKSQVSCSC RSFEYRGFLC RHAMLVLQIC 600 GISSIPTQYI LQRWTKDAKT SQLMVEGTEQ IRTRAQRYVA LSRQAIELSE EGSLTQESHN 660 IALRALVDAL KNCISINDAN KSAIEAGNET PGAHYVEGYQ GKMSTRRFKK KNTAKKQRDN 720 LGSDSISLNG YYGSHQNTQG MVQLNLMEPP HDSYYVDEES MQGLGQLNSM GQSHDEFFGA 780 QHSVHGLGHM DFRPSTSFSY SLEDEHNLRS TELHRDASR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
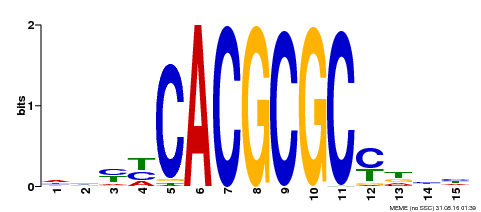
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010675660.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675661.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675662.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_010675663.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_019104685.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Refseq | XP_019104687.1 | 0.0 | PREDICTED: protein FAR-RED IMPAIRED RESPONSE 1 isoform X1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | F6HTN0 | 0.0 | F6HTN0_VITVI; Uncharacterized protein | ||||
| STRING | XP_010675660.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



