 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca39946.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 295aa MW: 33345.5 Da PI: 7.8097 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 143.3 | 1.3e-44 | 11 | 138 | 2 | 129 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k.kvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkge 98
ppG+rFhPtdeel ++yLk+k+ gk+l+ ++++e+d+y++ Pw+Lp k ++k+++ +wyf ++r++ky++g r+nr t +g+Wk++g+d++v +++
Dca39946.1 11 PPGIRFHPTDEELFMYYLKRKLLGKSLRH-QIMSEIDVYQFAPWELPaKsCLKTRDLNWYFICPRSRKYPSGGRTNRGTVHGFWKSSGNDRTVNY-RNR 107
9*************************999.89**************95357778888*************************************9.899 PP
NAM 99 lvglkktLvfykgrapkgektdWvmheyrle 129
vg ktL+fy+g+ pkge+t+Wv+heyrle
Dca39946.1 108 PVGKIKTLIFYRGKPPKGERTNWVIHEYRLE 138
*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.7E-54 | 4 | 160 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 51.933 | 10 | 160 | IPR003441 | NAC domain |
| Pfam | PF02365 | 7.8E-22 | 11 | 137 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 295 aa Download sequence Send to blast |
MDTALVSKSF PPGIRFHPTD EELFMYYLKR KLLGKSLRHQ IMSEIDVYQF APWELPAKSC 60 LKTRDLNWYF ICPRSRKYPS GGRTNRGTVH GFWKSSGNDR TVNYRNRPVG KIKTLIFYRG 120 KPPKGERTNW VIHEYRLEDK VLSEKGVALD SYVICKIFMK SGLGPKNGEN YGAPFMEEEW 180 DNSNDENVAS AQQQQQSVSV DIAGCSNGST ITSTLASPST LSTVQPYVAT TKGLPPIETE 240 EDELDRLLAK FTEECHNKPT PQSNVVDGLG NLAGRSSATS ANFDWSKFDW YDLCD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 2e-46 | 5 | 160 | 12 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 2e-46 | 5 | 160 | 12 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 2e-46 | 5 | 160 | 12 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 2e-46 | 5 | 160 | 12 | 165 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 3e-46 | 5 | 160 | 15 | 168 | NAC domain-containing protein 19 |
| 4dul_A | 2e-46 | 5 | 160 | 12 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 2e-46 | 5 | 160 | 12 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP). Transcriptional activator that acts as positive regulator of AOX1A during mitochondrial dysfunction. Binds directly to AOX1A promoter. Mediates mitochondrial retrograde signaling. {ECO:0000269|PubMed:24045017}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00583 | DAP | Transfer from AT5G64060 | Download |
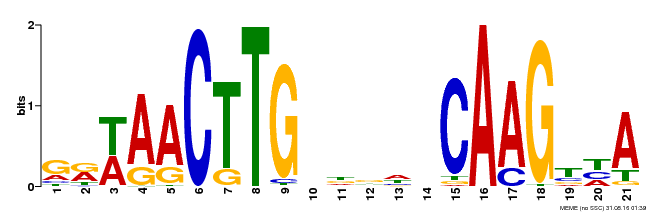
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold and drought stresses. {ECO:0000269|PubMed:17158162}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010674451.1 | 1e-106 | PREDICTED: NAC domain-containing protein 82 | ||||
| Swissprot | Q9XIC5 | 1e-66 | NAC17_ARATH; NAC domain-containing protein 17 | ||||
| TrEMBL | A0A0K9RG69 | 1e-97 | A0A0K9RG69_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010674451.1 | 1e-105 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64060.1 | 2e-76 | NAC domain containing protein 103 | ||||



