 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca40914.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 380aa MW: 41095.8 Da PI: 6.6194 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.9 | 5.9e-10 | 182 | 240 | 5 | 63 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklksev 63
kr++r NR +A rs +RK +i+eLe+kv++L++e ++L +l l+ l se+
Dca40914.1 182 KRAKRIWANRQSAARSKERKMRYIAELERKVQTLQTEATSLSAQLTLLQRDTNGLSSEN 240
9*********************************************9999888887776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 3.6E-15 | 178 | 242 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 10.955 | 180 | 243 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.74E-11 | 182 | 233 | No hit | No description |
| Pfam | PF00170 | 6.0E-9 | 182 | 240 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 1.6E-10 | 182 | 237 | No hit | No description |
| CDD | cd14703 | 1.26E-23 | 183 | 232 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 380 aa Download sequence Send to blast |
MEKEKSTGLP PTGKFPPFTP PPQTAAAAAS FSNDLSRMPE NPPKNTGHRR AHSEILTLPD 60 DISFDGDLGV VGGGGGVDGG ASFSDDTEED LVSMYIDMDK VNSATSSGFQ LGEASSSAAH 120 APVRGRHQHS QSMDGSTSLT PEMFVNGGGG GSGGGGEEVS PAEAKKAMSA AKLAELALID 180 PKRAKRIWAN RQSAARSKER KMRYIAELER KVQTLQTEAT SLSAQLTLLQ RDTNGLSSEN 240 NELKIRLQTM EQQVHLQDAL NDALKEEIQH LKMLTGQAMT NGGPMMNMPG FLGANQPFYP 300 NSQTMHTMLT AQQLQQLHIH SQKQPHQFPH HQLPLQQQQY HHPQEPSLQN TLLPDYKIRP 360 SGSAPSKEHA SDSLSGPSKD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor with an activatory role. | |||||
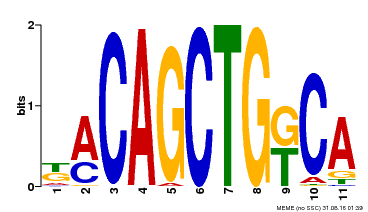
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00126 | DAP | Transfer from AT1G06070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010672248.1 | 1e-163 | PREDICTED: probable transcription factor PosF21 | ||||
| Swissprot | Q04088 | 1e-128 | POF21_ARATH; Probable transcription factor PosF21 | ||||
| TrEMBL | A0A2P5G0N3 | 1e-150 | A0A2P5G0N3_TREOI; Basic-leucine zipper transcription factor | ||||
| STRING | XP_010672248.1 | 1e-162 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06070.1 | 1e-121 | bZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



