 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca42443.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 147aa MW: 17463.8 Da PI: 6.6843 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 29.4 | 1.4e-09 | 90 | 124 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
k +yp++ ++ LA+ +gL++rq+ +WF N+R ++
Dca42443.1 90 KWPYPTEGDKIALAEMTGLDQRQINNWFINQRKRH 124
569*****************************985 PP
| |||||||
| 2 | ELK | 35.1 | 2.9e-12 | 44 | 65 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
+LK+qLlrK++++++sLk EFs
Dca42443.1 44 DLKDQLLRKFGSHISSLKLEFS 65
6********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03789 | 2.6E-9 | 44 | 65 | IPR005539 | ELK domain |
| SMART | SM01188 | 4.2E-6 | 44 | 65 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 9.935 | 44 | 64 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.552 | 64 | 127 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 2.1E-20 | 65 | 139 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 2.1E-12 | 66 | 131 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-28 | 69 | 128 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.53E-11 | 73 | 128 | No hit | No description |
| Pfam | PF05920 | 2.7E-18 | 84 | 123 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 102 | 125 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010073 | Biological Process | meristem maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 147 aa Download sequence Send to blast |
MFISFKPHVI YFMFAEEAAA SSDEECSGGE VDVHDMQPKD EERDLKDQLL RKFGSHISSL 60 KLEFSKKKKK GKLPREARQM LFEWWNAHYK WPYPTEGDKI ALAEMTGLDQ RQINNWFINQ 120 RKRHWKPSEN MQFALMDNIS GQYFPDD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Plays a role in meristem function. Contributes to the shoot apical meristem (SAM) maintenance and organ separation by controlling boundary establishment in embryo in a CUC1, CUC2 and STM-dependent manner. Involved in maintaining cells in an undifferentiated, meristematic state. Probably binds to the DNA sequence 5'-TGAC-3'. {ECO:0000269|PubMed:16798887}. | |||||
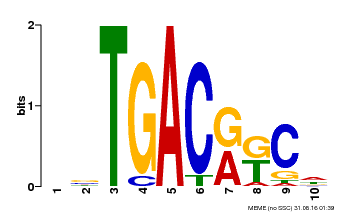
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00670 | PBM | Transfer from GRMZM2G087741 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Seems to be repressed by AS2 and AS1 but induced by STM, CUC1 and CUC2. {ECO:0000269|PubMed:11311158, ECO:0000269|PubMed:16798887}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021727071.1 | 2e-78 | homeobox protein knotted-1-like 6 isoform X2 | ||||
| Swissprot | Q84JS6 | 1e-60 | KNAT6_ARATH; Homeobox protein knotted-1-like 6 | ||||
| TrEMBL | A0A0K9RP31 | 2e-76 | A0A0K9RP31_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010695813.1 | 5e-77 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G23380.1 | 3e-51 | KNOTTED1-like homeobox gene 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



