 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca4257.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 508aa MW: 56671.9 Da PI: 7.9284 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 99.2 | 2.6e-31 | 201 | 258 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+Dgy+WrKYGqK+vkgse+prsYY+C++++Cp+kkkvers+ +++v+ei+Y+g+Hnh+k
Dca4257.1 201 EDGYCWRKYGQKQVKGSENPRSYYKCSYPNCPTKKKVERSR-EGHVTEIVYKGSHNHPK 258
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 106 | 2e-33 | 357 | 415 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+CtsagCpv+k++er+++d ++v++tYeg+Hnh+
Dca4257.1 357 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSAGCPVRKHIERASHDMRAVITTYEGKHNHD 415
59********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 7.6E-27 | 192 | 259 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 23.02 | 195 | 259 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.28E-24 | 198 | 259 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.1E-33 | 200 | 258 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.4E-23 | 201 | 257 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 3.1E-37 | 342 | 417 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 5.36E-29 | 349 | 417 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.744 | 352 | 417 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.3E-38 | 357 | 416 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.4E-25 | 358 | 415 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0009788 | Biological Process | negative regulation of abscisic acid-activated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0010508 | Biological Process | positive regulation of autophagy | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0070370 | Biological Process | cellular heat acclimation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 508 aa Download sequence Send to blast |
MTSPFTDFFA SNNNNNNNNN DDNNNGIERS NSISSWGGFN ELVEAPKFKS YAPSSISVSK 60 PNSFATTGPN SFSPSIFLDS PLLNVQNFPS PTVGAFGGEN FNWGASNNND THKVVKDEAK 120 NINNNNFTFH APNITTDGWK LSESKSQNDK INNDNSSSQS GYNSNQVIPW EMSSQKSDDN 180 VQRSYSSQPA VPMNRAQKRA EDGYCWRKYG QKQVKGSENP RSYYKCSYPN CPTKKKVERS 240 REGHVTEIVY KGSHNHPKPQ PGKKTSDVVS NTLVVHGMTE NNNSTFTPET SSVSMDDDEE 300 FDQINSAISV TRKDDEIEPQ PKRWKGENEA EVMSGYGGRT VKEPRVIVQT TSEIDILDDG 360 YRWRKYGQKV VKGNPNPRSY YKCTSAGCPV RKHIERASHD MRAVITTYEG KHNHDVPAAR 420 GSSGYNNVPV PVRPLSAVPN QTNHFNSYKP FNNNTTNGIK PETQSQSQSP FTLQMLQSPG 480 SFGFSGFGNQ NEAKEEPMDN KFLDTYFG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-37 | 201 | 418 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-37 | 201 | 418 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00299 | DAP | Transfer from AT2G38470 | Download |
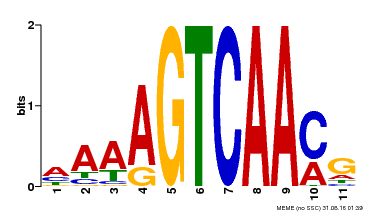
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019106270.1 | 0.0 | PREDICTED: probable WRKY transcription factor 33 | ||||
| TrEMBL | A0A0K9Q8X6 | 0.0 | A0A0K9Q8X6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010683807.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38470.1 | 1e-109 | WRKY DNA-binding protein 33 | ||||



