 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca47961.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 283aa MW: 31725.1 Da PI: 9.3244 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.6 | 1.1e-11 | 5 | 57 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqkyl 48
+++WT+eE++ l ++v+++G g Wk I + R+ ++k++w++++
Dca47961.1 5 KQKWTAEEEQALREGVHKYGAGKWKFIHSDPVfgnvlHSRSNIDLKDKWRNLR 57
79****************************9999***9*************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 15.422 | 1 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.52E-16 | 2 | 58 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.8E-8 | 4 | 59 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.2E-14 | 5 | 56 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 9.4E-9 | 5 | 57 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 6.13E-20 | 6 | 57 | No hit | No description |
| SMART | SM00526 | 7.8E-6 | 96 | 187 | IPR005818 | Linker histone H1/H5, domain H15 |
| PROSITE profile | PS51504 | 15.813 | 122 | 192 | IPR005818 | Linker histone H1/H5, domain H15 |
| SuperFamily | SSF46785 | 7.62E-11 | 122 | 201 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 5.2E-8 | 125 | 196 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF00538 | 4.5E-5 | 128 | 185 | IPR005818 | Linker histone H1/H5, domain H15 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006334 | Biological Process | nucleosome assembly | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0000786 | Cellular Component | nucleosome | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 283 aa Download sequence Send to blast |
MGNPKQKWTA EEEQALREGV HKYGAGKWKF IHSDPVFGNV LHSRSNIDLK DKWRNLRAAS 60 EGTGTRSRTP KPKLTEDAPV GLAPSTNVAA PVVIPVAPKP VTNPVKEEHV KETPDAKNVS 120 RSSNDMVGMI FEALSASTEQ NGLDIGSIVN YIEQRITDEL PQNFRKQLGA RLRRLVLHER 180 LEKVENCYRL KKNSLAETNA VVRKDFPRQP LIPAQVRINE SLQEAAKTAA YKVAEAEEKT 240 HLAALSVREA ERFVKMQEES DAVLQLAQEI FDRLHNNEAL LIT |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
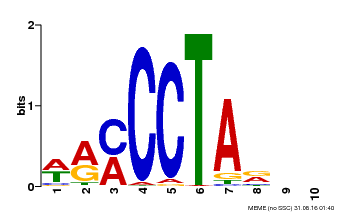
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010696540.1 | 1e-101 | PREDICTED: telomere repeat-binding factor 4 | ||||
| TrEMBL | A0A2P2L4C7 | 5e-77 | A0A2P2L4C7_RHIMU; Uncharacterized protein | ||||
| STRING | XP_010696540.1 | 1e-101 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G17520.1 | 6e-33 | MYB_related family protein | ||||



