 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca50574.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 568aa MW: 64002.2 Da PI: 4.9498 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 75 | 1e-23 | 187 | 242 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLG.GsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k+r++Wt eLH++Fv+av+q+G s+k Pk+il+lm+v+ Lt+e+v+SHLQkYRl
Dca50574.1 187 KARVVWTIELHQKFVKAVNQIGiDSDKVGPKKILDLMNVPWLTRENVASHLQKYRL 242
68*******************7447899***************************8 PP
| |||||||
| 2 | Response_reg | 78.7 | 2e-26 | 22 | 130 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTES CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGak 95
vl+vdD+p+ +++l+++l+k y ev++ ++eal+ll++++ +D+++ D++mp+mdG++ll++ e +lp+i+++ ge + + + ++ Ga
Dca50574.1 22 VLVVDDDPTWLKILEKMLKKCSY-EVTTSGLAREALRLLRDRKdgFDIVISDVNMPDMDGFKLLEHVGLEM-DLPVIMMSVDGETSRVMKGVQHGAC 116
89*********************.***************888889**********************6644.8************************ PP
EEEESS--HHHHHH CS
Response_reg 96 dflsKpfdpeelvk 109
d+l Kp+ ++el +
Dca50574.1 117 DYLLKPIRMKELKN 130
**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.40.50.2300 | 1.1E-43 | 19 | 169 | No hit | No description |
| SuperFamily | SSF52172 | 5.83E-35 | 19 | 143 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 9.5E-31 | 20 | 132 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 42.913 | 21 | 136 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.7E-23 | 22 | 130 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 7.18E-30 | 23 | 135 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-27 | 185 | 247 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.81E-17 | 186 | 246 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.5E-21 | 187 | 242 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.3E-6 | 189 | 241 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 568 aa Download sequence Send to blast |
MIMESGFSSP RRSEAFPAGL RVLVVDDDPT WLKILEKMLK KCSYEVTTSG LAREALRLLR 60 DRKDGFDIVI SDVNMPDMDG FKLLEHVGLE MDLPVIMMSV DGETSRVMKG VQHGACDYLL 120 KPIRMKELKN IWQHVVRKRL HEVRDIEFID DRDHPNDGYY GGDPTTLKKR KDVENDKESN 180 DLSLNKKARV VWTIELHQKF VKAVNQIGID SDKVGPKKIL DLMNVPWLTR ENVASHLQKY 240 RLYLSRIGKG DDCMESSFGA TKNTDFSPKE SGGGSISVQN SSYYALHSDV TSNPMMPPLQ 300 SSECKTHDQK LKTTEPKKSV NNENSDPQRT ANSKAGLNST FQHTVFDSTV SGQWQFKPEL 360 KPQPKSKNCF NQPVVPILQQ QIPSDLLEHA PEINPGLFNE SDGFKLIDVK PLISKPYSVP 420 NPGTFGCIFD VPLDVKNQNC CSPSKNFDKQ DPLAVLESTT FDPQVLETTT FDPNSNNVKG 480 SENESQNQAT SETEPAFGVV SEDLMFRWLQ GGDGFGIDDV GFSDSNFAEP INDQVPFPPY 540 YMPKFGVEYE YPYDWVEYPV VDQGLLFS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-17 | 186 | 247 | 4 | 63 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
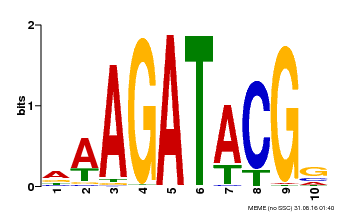
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins (By similarity). {ECO:0000250, ECO:0000269|PubMed:12610214}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00010 | PBM | Transfer from AT1G67710 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010682390.1 | 0.0 | PREDICTED: two-component response regulator ARR11 isoform X1 | ||||
| Swissprot | Q9FXD6 | 1e-122 | ARR11_ARATH; Two-component response regulator ARR11 | ||||
| TrEMBL | A0A0K9QRY8 | 0.0 | A0A0K9QRY8_SPIOL; Two-component response regulator | ||||
| STRING | XP_010682390.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G67710.1 | 1e-113 | response regulator 11 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



