 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca53252.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 836aa MW: 91916.5 Da PI: 6.3669 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.4 | 1.2e-18 | 16 | 74 | 3 | 57 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
k ++t+eq+e+Le+l++++++ps +r++L + + +++ +q+kvWFqNrR +ek+
Dca53252.1 16 KYVRYTPEQVEALERLYHECPKPSSLRRQQLIRDCpilsNIEPKQIKVWFQNRRCREKQ 74
5679*****************************************************97 PP
| |||||||
| 2 | START | 175.6 | 3e-55 | 160 | 368 | 2 | 205 |
HHHHHHHHHHHHHHHC-TT-EEEEEXCCTTEEEEEEESSS.SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-SEEEEEEEECTT..EEEEEEEEXX CS
START 2 laeeaaqelvkkalaeepgWvkssesengdevlqkfeeskvdsgealrasgvvdmvlallveellddkeqWdetlakaetlevissg..galqlmvael 98
+aee+++e+++ka+ ++ Wv+++ +++g++++ +++ s++++g a+ra+g+v +++ v+e+l+d++ W +++++++l+v+ ++ g+++l +++l
Dca53252.1 160 IAEETLAEFLSKATGTAVEWVQMPGMKPGPDSIGIVAISHGCPGVAARACGLVGLEPT-RVAEILKDRPSWFCDCRAVDVLNVLPTAngGTIELLYMQL 257
7899******************************************************.8888888888****************9999********** PP
TTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--....-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXXHHHHHHHHHHHHHHHH CS
START 99 qalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppe...sssvvRaellpSgiliepksnghskvtwvehvdlkgrlphwllrslvksglaega 193
+a+++l+p Rdf+ +Ry+ +++g++v++++S++++q+ p+ +++vRae+lpSg+li+p+++g+s +++v+h +l+ ++++++lr+l++s+++ ++
Dca53252.1 258 YAPTTLAPaRDFWLLRYTSVMEDGSLVVCERSLNNTQNGPSmpaVQNFVRAEMLPSGYLIRPCEGGGSIIHIVDHLNLEPWSVPEVLRPLYESSTVVAQ 356
****************************************999899***************************************************** PP
HHHHHHTXXXXX CS
START 194 ktwvatlqrqce 205
kt++a+l+++++
Dca53252.1 357 KTTMAALRQLRQ 368
********9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50071 | 15.354 | 11 | 75 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.0E-15 | 13 | 79 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.45E-16 | 16 | 79 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 6.64E-16 | 16 | 76 | No hit | No description |
| Pfam | PF00046 | 3.0E-16 | 17 | 74 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 2.3E-18 | 18 | 74 | IPR009057 | Homeodomain-like |
| CDD | cd14686 | 1.28E-6 | 68 | 107 | No hit | No description |
| PROSITE profile | PS50848 | 24.946 | 150 | 365 | IPR002913 | START domain |
| CDD | cd08875 | 1.16E-78 | 154 | 370 | No hit | No description |
| SuperFamily | SSF55961 | 1.65E-37 | 159 | 371 | No hit | No description |
| Gene3D | G3DSA:3.30.530.20 | 3.3E-22 | 159 | 366 | IPR023393 | START-like domain |
| SMART | SM00234 | 2.3E-41 | 159 | 369 | IPR002913 | START domain |
| Pfam | PF01852 | 1.1E-52 | 160 | 368 | IPR002913 | START domain |
| Pfam | PF08670 | 5.5E-51 | 693 | 835 | IPR013978 | MEKHLA |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010075 | Biological Process | regulation of meristem growth | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0048263 | Biological Process | determination of dorsal identity | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 836 aa Download sequence Send to blast |
MPMSCKDGKG VMDNGKYVRY TPEQVEALER LYHECPKPSS LRRQQLIRDC PILSNIEPKQ 60 IKVWFQNRRC REKQRKEASR LQAVNRKLTA MNKLLMEEND RLQKQVSHLV YENGYFRQQT 120 QKAGIASKDT SCESVVSSGQ QHLTPQHPPR DASPAGLLSI AEETLAEFLS KATGTAVEWV 180 QMPGMKPGPD SIGIVAISHG CPGVAARACG LVGLEPTRVA EILKDRPSWF CDCRAVDVLN 240 VLPTANGGTI ELLYMQLYAP TTLAPARDFW LLRYTSVMED GSLVVCERSL NNTQNGPSMP 300 AVQNFVRAEM LPSGYLIRPC EGGGSIIHIV DHLNLEPWSV PEVLRPLYES STVVAQKTTM 360 AALRQLRQIA QEISQPTVSN WGRRPAALRD LSQRLNRGFN EALNGFTDEG WTMIGNDGID 420 DVTVLVNSSP DKLMALNLTY ANGFSAVSNT VLCAKASMLL QNVPPAVLLR FLREHRSEWA 480 DNNIDTYLAA ATKISPCSLP GTRIGGFGNQ VILPLAHTIE HEELLEVIKL EGVVSCPEDA 540 MMPRDMFLLQ MCSGMDENAV GTCAELIFAP IDASFADDAP LLPSGFRIIP LDSAKEASSP 600 NRTLDLASSL EIGPSVSKSA GNQSASATNM RSVMTIAFQF AFESHLQDNV TSMARQYVRS 660 IISSVQRVAL ALSPHMGSQA GLRSPRGNPE AHTLASWICQ SYRCFLGVEL LKSAGEGGDA 720 VLKSLWHHSD AIMCCTLKAL PVFTFANQAG LDMLETTLVA LQDITLEKIL DEHGRKTLCS 780 EFPQILQQGF ACLQGGICLS SMGRPISYER AVAWKVLNEE ENAHCICFMF INWSFV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of meristem development to promote lateral organ formation. May regulates procambial and vascular tissue formation or maintenance, and vascular development in inflorescence stems. {ECO:0000269|PubMed:15598805, ECO:0000269|PubMed:15705957, ECO:0000269|PubMed:16617092}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00200 | DAP | Transfer from AT1G52150 | Download |
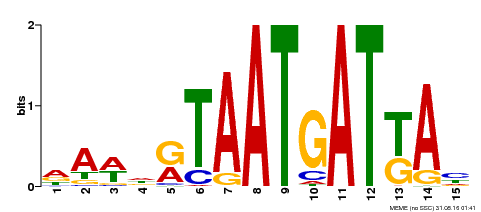
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. Repressed by miR165 and miR166. {ECO:0000269|PubMed:15773855, ECO:0000269|PubMed:16033795, ECO:0000269|PubMed:16617092, ECO:0000269|PubMed:17237362}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010691423.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ATHB-15 isoform X1 | ||||
| Swissprot | Q9ZU11 | 0.0 | ATB15_ARATH; Homeobox-leucine zipper protein ATHB-15 | ||||
| TrEMBL | A0A0K9RV39 | 0.0 | A0A0K9RV39_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010691422.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52150.1 | 0.0 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



