 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca54358.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 845aa MW: 96708.5 Da PI: 7.3088 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 81.4 | 1.5e-25 | 87 | 188 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHe 88
fY++YA+++GF++ +++s++sk+++e+++++f Csk+g+++e +k +++ +r+ +t+Cka+++vk++ dg+w+++++ +eHnH+
Dca54358.1 87 FYQDYARSMGFNTAIQNSRRSKTSREFIDAKFACSKYGTKREYDKGnrprvrqisnqePDSATGRRSCGKTNCKASMHVKRRMDGRWVIHSFIKEHNHD 185
9**************************************999999889999988775545555589999****************************** PP
FAR1 89 lap 91
l p
Dca54358.1 186 LLP 188
975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 3.0E-23 | 87 | 188 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.6E-28 | 286 | 378 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.248 | 567 | 603 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 2.4E-4 | 577 | 602 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 5.9E-9 | 578 | 605 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 845 aa Download sequence Send to blast |
MDIDLRLPSG EQDKHDEETH VLSNMLGSDD NLHDEEARTA LEAVVLHGNE EVTLACATTD 60 LVDFKDDVNL EPLPGMEFVS HQEAYAFYQD YARSMGFNTA IQNSRRSKTS REFIDAKFAC 120 SKYGTKREYD KGNRPRVRQI SNQEPDSATG RRSCGKTNCK ASMHVKRRMD GRWVIHSFIK 180 EHNHDLLPAQ AVSEQTRKIY AAMARHYAEY KSVIGLKNEL KSPFEKTRNL VLDPGDAKLA 240 LNFCKQMQVI NSHFFYAVEV SDDLRLKTLL WVDAKSRHDY VNFTDVISVD TTYIRNKYKI 300 PLALLVGVNQ HYQFMLLGCA IVTDESPTTY SWILQTWLQA MGGQAPKVII TDQENSFKSA 360 VSEVFPHARH CYHLWHVLGK VSENLGQVIK THDNFMTKFE KCIYRSWTDD EFEKRWQKLV 420 ARFGLDKDSD WIHSLYEDRR LWVPLYMKDT LLAGLSTVPR SESVNSYFDR YVHKRTTISE 480 FLKQYESIIQ DRYEEEARAD ADTWNKQPML KSPSPLEKHM STIYTQAVFK KFQVEILGAV 540 ACHPKRERQD DQMTTFKVRD LEKDPDFMVT WNEMKSEVSC ICRLFEYKGF LCRHAMIVLQ 600 MCGLSMIPSL YILRRWTKEA KERHLTGESS MQVSSRVQRY NDLCNRAVKL SEEGSLSQES 660 YSIAFLALDE AFSSCIAANT ACKTLSEVGT SLTHELLSIE NGSPSQSLGK TSKKKSLTKK 720 RKVNSEGEPI TAAPQDTLQP LDKLSSRPVT SLEGLYGTQQ NVQGMVQLNL MGPMSENYYG 780 SQPAIQGLGQ LNSIASGHDG YYSQQGMHGL GQMDFFRAPT NFGYGIRDEP IVRSTQLHDD 840 RHKHS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
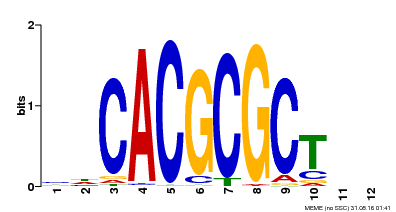
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010680157.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X2 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A0K9QP72 | 0.0 | A0A0K9QP72_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010680156.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



