 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca56299.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 410aa MW: 45685.5 Da PI: 4.9687 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.2 | 3e-16 | 50 | 94 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT++E+ ++++a k++G W++I +++g ++t+ q++s+ qk+
Dca56299.1 50 RERWTEDEHTKFLEALKLHGRA-WRKIEEHIG-TKTAVQIRSHAQKF 94
78******************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.73E-16 | 44 | 98 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.728 | 45 | 99 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-9 | 47 | 97 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.7E-16 | 48 | 97 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.2E-12 | 49 | 97 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-13 | 50 | 93 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.19E-10 | 52 | 95 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 410 aa Download sequence Send to blast |
MVSQEKTSAD NSNSVLSSEN VVSMDVCVQS DIDEGQIPKI RKPYTITKQR ERWTEDEHTK 60 FLEALKLHGR AWRKIEEHIG TKTAVQIRSH AQKFFSKVVR ETINSNASTT GPIEIPPPRP 120 KRKPSHPYPR KLALPTKKDN SPDQQGRSIS PYSSVDDQQN QSPTSVLSAL ASDTLASDSD 180 TPNDSPSPDC FNDKTDVLPS DDNSPQDDVR ETPLTPDERV SVKLELFPSM DSFDKPQATA 240 TKVLKLFGKD VVVSSTSSCL NKSPSMDMGE SVLNETAENS QDCQTLYCVD SSGNHVEKPV 300 PLPWWVFFKG MPVSPLETQV ENPQEKETER EGSFAGSNSG SVNAEGNTDN NYDTESQSNY 360 EIKSLLEKRS SINKPNKGFV PYKRCFSERD MQSSTASINE DESRRIRLCL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
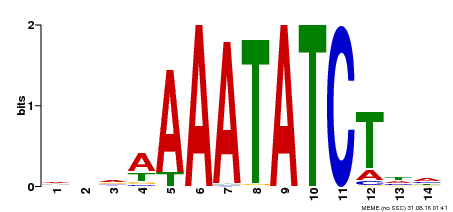
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021856034.1 | 1e-143 | protein REVEILLE 1 isoform X1 | ||||
| TrEMBL | A0A0K9R483 | 1e-142 | A0A0K9R483_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010675239.1 | 1e-138 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 2e-43 | MYB_related family protein | ||||



