 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca58184.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 694aa MW: 77442.2 Da PI: 4.9461 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 37.6 | 5.2e-12 | 640 | 689 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqk 46
r++W+ E++ l ++v+++G++ Wk+I +gRt+ ++k++w++
Dca58184.1 640 RKKWSSLEEDMLREGVQKYGKR-WKLILTDSKyvkifEGRTDVDLKDKWRN 689
789*******************.***************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.899 | 635 | 694 | IPR017930 | Myb domain |
| SMART | SM00717 | 6.5E-8 | 639 | 693 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.8E-9 | 640 | 689 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-12 | 640 | 692 | IPR009057 | Homeodomain-like |
| CDD | cd11660 | 6.00E-14 | 641 | 692 | No hit | No description |
| SuperFamily | SSF46689 | 1.79E-10 | 642 | 692 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 694 aa Download sequence Send to blast |
MEETIAKWII EYFLRRRSDD KVINALISSL PLAVDDYRLK KTMLLHRIET EASKGWISEK 60 IVELIEIIEA LDHEEGKMAS ELMKTAYCLV AVDCTVRFLV QTGEKNEKYL EAVENLWRNR 120 VCKSAGLVSE MSKRWFDDIE AAGNDSAVRE KLWMRNTRNE AMKAVIEYVK EQWECLGPTF 180 LEVVAAKVGD DVEGTWLNDG DRGVYGGGSQ QGQPVVRRKH LAVKTKRPRG ARLSDVEEEA 240 VNTTGDGITE TEGAKFPSLS SEMDKAKEAI CTGFAAETRD IRGEKRSDVD GDTMNKAGDS 300 TAETVRAEDR TPEINRAKET SSSGFTQGPS VGGRVVDGVR TAQTLTFMTP LHRGVDGDVE 360 AEQNRRVIDD SLEVKMQQRE LRERTRELLA AVCDPLPEAL KTAQTCISLT ETNNHETAPQ 420 NRVEPDMPTS AEAVRVQTAQ EPAETTISNS ENNAEVSKQA GREVDPPRGS AEDGSGNVKE 480 TARHDDADED VTVNLVAQIN LRKRKNVDAP SDPNEKGSVD GTSCTLNEDA QGNVTQIPEA 540 QNNVHKRAKV DAASRGSDEN VAVHEDALGN AAQDPPGAQD NARKRSLMEP NDTSHTFEWD 600 DDIENSAGSP VRPTLPTPNV QRVSPLRVPE YRNLLRRRER KKWSSLEEDM LREGVQKYGK 660 RWKLILTDSK YVKIFEGRTD VDLKDKWRNM SRHS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
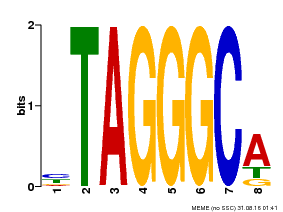
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 4e-19 | TRF-like 5 | ||||



