 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Dca7798.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Caryophyllaceae; Caryophylleae; Dianthus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 982aa MW: 108702 Da PI: 5.9182 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 128.5 | 2.4e-40 | 135 | 212 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC+adl++a++y+rrhkvCevhskap+++v ++ qrfCqqCsrfh ++efD +krsCr+rLa+hn+rrrk+++
Dca7798.1 135 MCQVEGCTADLTNARDYNRRHKVCEVHSKAPEAIVGNVVQRFCQQCSRFHLIQEFDGDKRSCRKRLAGHNKRRRKNNP 212
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.9E-32 | 130 | 197 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.24 | 133 | 210 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.14E-37 | 134 | 214 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.7E-30 | 136 | 209 | IPR004333 | Transcription factor, SBP-box |
| SMART | SM00248 | 260 | 745 | 775 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF48403 | 9.7E-9 | 746 | 861 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.9E-9 | 748 | 865 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.96E-10 | 749 | 862 | No hit | No description |
| SMART | SM00248 | 1.9 | 803 | 833 | IPR002110 | Ankyrin repeat |
| Pfam | PF00023 | 0.018 | 804 | 827 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 982 aa Download sequence Send to blast |
MKAVGKRSLE WDLNDWKWDS NLLLATPLNP GAYDRSRSSH FSPLVPRDLH LAANTNVGVS 60 NSLLPSLEET NPYNDGRIQR TAEKKRTIVV VHEDEGLNDE VTPVNLKLGE QVYPIVDAEG 120 DKGRKSKNGG TCHPMCQVEG CTADLTNARD YNRRHKVCEV HSKAPEAIVG NVVQRFCQQC 180 SRFHLIQEFD GDKRSCRKRL AGHNKRRRKN NPETSPSNVG SVADGINAGQ ILNLLLRLLS 240 NVPTNGSTQE NDQDLLSQIL RNLAASSGGC NTSGLLLGSQ ASPDVGTSNG IPEKDHSNNQ 300 ELSQTTAYAQ TGTLTRENQS KVSQTTSVPQ PKVFQATSDR VLTNGCVQDA SFEKQKLSGI 360 DLNAVYHDDS EVCIDDQTNV AFVNGAYWAE HTLPSPPPTS GNSSSTSGRS SSSSGEAQCR 420 TDRIILKLFE KDPNDMPNVL RSQILDWLSH SPTDIEGYIR PGCIVLTLYL RLNTSLWEQF 480 CYDLRSSLSR LLEASNDPFW KTGWIYTRIQ QRAAFVYDGQ VVLDTPLPFR SLRSQISSIS 540 PIALPTGETV KLVVRGSNLS GPTSRLLCGV EGKYLVQENC CDLVEGTEVT TEQDEIESLS 600 FSCSIPTVLG RGFIEVEDYD LSGCFFPFIV AEPELCYEIC TLEQEMQTVI EDEYNNQSEN 660 GEADAKSKAL DFVHEMGWLL HTYSLRFTEN QQVPLVNLFS FERLKWLVEF SMDHDWPSVL 720 RKLLDLLFSG IVDIGNYASV EDGLADIAPL HAAVRRNSRL MVEFLLRYVP NKVGNAVVGL 780 KQNQPDYGAS GNMLFKPDVV GAGGLTPLHI AASSTGSENV LDALLEDPGM VGIEAWKNAR 840 DSTGLTPSDY ASLRGHYRYL HLVQRKTNKK SANKQSTEKH VVVDIPSLSL LKPNQIPSDK 900 LKPIEFNSFY TEKPQTGQSC KLCDQIPYHR FGSGPLAFRP VMMSLVAIAV VCVCTALLFK 960 SEPQVCGIFG RFRWEALKYG AM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 2e-30 | 135 | 209 | 10 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
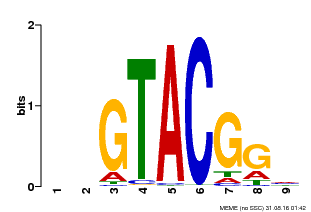
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010672614.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Refseq | XP_010672615.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A0J8CTR7 | 0.0 | A0A0J8CTR7_BETVU; Uncharacterized protein | ||||
| STRING | XP_010672614.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||



