 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do000373.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 304aa MW: 32122 Da PI: 7.0851 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 56.5 | 7e-18 | 58 | 106 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+ GVr+++ +gr++AeIrdp++ + r++lg+f+ta+eAa a+++a+++++g
Do000373.1 58 FLGVRRRP-WGRYAAEIRDPTT---KERHWLGTFDTAQEAALAYDRAALSMKG 106
56******.**********965...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 20.547 | 57 | 114 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.24E-25 | 57 | 113 | No hit | No description |
| SMART | SM00380 | 1.4E-29 | 57 | 120 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-7 | 58 | 69 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.51E-20 | 58 | 115 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 4.9E-28 | 58 | 114 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 2.6E-11 | 60 | 106 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-7 | 80 | 96 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010102 | Biological Process | lateral root morphogenesis | ||||
| GO:0010432 | Biological Process | bract development | ||||
| GO:0010451 | Biological Process | floral meristem growth | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 304 aa Download sequence Send to blast |
MNTRGSGSSG GGGNQTLMAF SEQPKPASAQ PQPSPPSSPS ERPPAGRGRR RAQEPGRFLG 60 VRRRPWGRYA AEIRDPTTKE RHWLGTFDTA QEAALAYDRA ALSMKGAQAR TNFVYTHAAY 120 NYPPFLAPFH AQGSSYGHAP SSMQYGQQHG GVGAAHIGSY HHHHHHYQAS AGSGGGASSS 180 SSVPAAVDRA DGTLLMDHRN GHDFLFASAD DNSGYLSSVV PESCLRPRSS AAAVEDLRRY 240 SDADAYGMGM GLREDVDDLA QMVAGFWGGA GDAEQLCGFP SGGGDMVASS QGSDGYSPFS 300 FLSH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 3e-18 | 48 | 113 | 5 | 71 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Required to prevent the formation of axillary meristems within the spikelet meristem and permit the subsequent establishment of floral meristem identity (PubMed:12835399, PubMed:14503923). Mediates the transition from spikelet to floret meristem (PubMed:14503923). Determines the transition from panicle branching to spikelet formation. May specify floral organ identity by regulating the class B genes (Agamous-like genes) MADS6 and MADS17, as well as class E genes MADS1, MADS7 and MADS8 in floral meristem (PubMed:26744119). Possesses transactivation activity (PubMed:12835399). {ECO:0000269|PubMed:12835399, ECO:0000269|PubMed:14503923, ECO:0000269|PubMed:26744119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00520 | DAP | Transfer from AT5G18560 | Download |
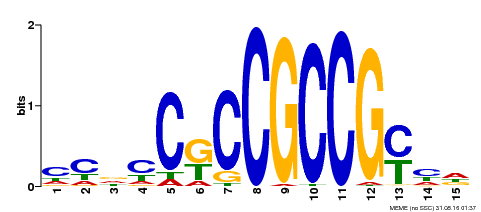
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do000373.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT036365 | 1e-137 | BT036365.1 Zea mays full-length cDNA clone ZM_BFb0108M21 mRNA, complete cds. | |||
| GenBank | KJ727547 | 1e-137 | KJ727547.1 Zea mays clone pUT5407 AP2-EREBP transcription factor (EREB151) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004958652.1 | 1e-154 | ethylene-responsive transcription factor ERF086 | ||||
| Swissprot | Q8H3Q1 | 1e-115 | FZP_ORYSJ; Ethylene-responsive transcription factor FZP | ||||
| TrEMBL | A0A1E5UMV0 | 0.0 | A0A1E5UMV0_9POAL; Ethylene-responsive transcription factor ERF086 | ||||
| STRING | Si032771m | 1e-154 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP10525 | 28 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18560.1 | 2e-35 | ERF family protein | ||||



