 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do001153.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 470aa MW: 49318.7 Da PI: 9.1531 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 47.9 | 3.2e-15 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+ lv +v+ G+g+W++++ g+ R+ k+c++rw +yl
Do001153.1 14 KGPWTPEEDLVLVSYVQDKGPGNWRAVPTNTGLMRCSKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 50.5 | 4.7e-16 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+ E++l+v++ ++lG++ W++Ia++++ Rt++++k++w+++l
Do001153.1 67 RGNFTEKEEQLIVHLQALLGNR-WAAIASYLP-ERTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.449 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.73E-29 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-11 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.0E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.03E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.618 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-16 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.9E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.02E-12 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010468 | Biological Process | regulation of gene expression | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 470 aa Download sequence Send to blast |
MGRPPCCEKT GVKKGPWTPE EDLVLVSYVQ DKGPGNWRAV PTNTGLMRCS KSCRLRWTNY 60 LRPGIKRGNF TEKEEQLIVH LQALLGNRWA AIASYLPERT DNDIKNYWNT HLKKKLSKVG 120 TGEGGAGAGT SGEAKSGRSA AHKGLWERRL QTDIHTARQA LREALSLDPA LPPAKLEPMP 180 APLPLTQPPV PLSPATYASS TENIRRLLQG WMRPGAGKAS SWPRSSASVL SGGEGASGSH 240 SGTARTPEGS TGTSKLEDAG KASPVPPFSM LESWLLDDGM GHGSAGLMGV PLADPKVGTG 300 EGGAGAGTSG EAKSGRSAAP KGQWERRLQT DIHTARQALR EALSLDTALP LAKLEPMPAP 360 QPQPPAMYAS STENIAHLLE GWMRPGAGKA SSWSRSSASV LSGGEGGPGS HSGTARTPEG 420 STGTGKVEDM GAAGPVPPFL MLESWLLDDG MGHGDAGLMG VPLADPCQFF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 6e-25 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00579 | DAP | Transfer from AT5G62470 | Download |
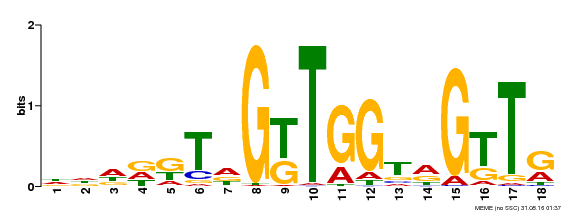
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do001153.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025821217.1 | 1e-139 | myb-related protein 306-like isoform X1 | ||||
| Swissprot | P81392 | 3e-99 | MYB06_ANTMA; Myb-related protein 306 | ||||
| TrEMBL | A0A3L6RLD1 | 1e-148 | A0A3L6RLD1_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.Fa00895.1.p | 1e-148 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP229 | 38 | 296 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G28910.1 | 1e-90 | myb domain protein 30 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



