 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do004107.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 289aa MW: 32971.1 Da PI: 8.9979 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.5 | 1.3e-31 | 63 | 113 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fss+g+lyeys+
Do004107.1 63 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSN 113
79***********************************************95 PP
| |||||||
| 2 | K-box | 98.4 | 1.1e-32 | 131 | 228 | 4 | 100 |
K-box 4 ssgks.leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkklee 100
ss ++e + ++++qe+a+L+++i nLq+++R l+G++++++s+k+L+qLe +L+k+l kiR++Kne+l++++e++q++e elq++n +Lr+++ee
Do004107.1 131 SSAAGtVAEVTIQHYKQESARLRQQITNLQNSNRTLIGDSITTMSHKDLKQLEARLDKGLVKIRARKNEVLCSELEYMQRREMELQNDNLYLRSRVEE 228
44444689999***********************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.7E-40 | 55 | 114 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.221 | 55 | 115 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 3.53E-33 | 56 | 128 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 3.64E-45 | 56 | 129 | No hit | No description |
| PRINTS | PR00404 | 9.4E-33 | 57 | 77 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 57 | 111 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.2E-26 | 64 | 111 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 9.4E-33 | 77 | 92 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 9.4E-33 | 92 | 113 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 14.019 | 142 | 232 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 7.0E-24 | 142 | 226 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0090376 | Biological Process | seed trichome differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MHIQEEQQAT PSTDIMWGFS GQSTLTSTGL KLKEPEPLLS SPGSGSAGAA AAEKNGRGKI 60 EIKRIENTTN RQVTFCKRRN GLLKKAYELS VLCDAEVALI VFSSRGRLYE YSNNSVKATI 120 ERYKKATSDN SSAAGTVAEV TIQHYKQESA RLRQQITNLQ NSNRTLIGDS ITTMSHKDLK 180 QLEARLDKGL VKIRARKNEV LCSELEYMQR REMELQNDNL YLRSRVEENE RAQQTVNMME 240 APSTSEYQQG FIPYDPIRSF LQFNIMQQQP QFYSQQEDRK DFNFNFNLG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6bz1_A | 2e-20 | 56 | 143 | 2 | 89 | MEF2 CHIMERA |
| 6bz1_B | 2e-20 | 56 | 143 | 2 | 89 | MEF2 CHIMERA |
| 6bz1_C | 2e-20 | 56 | 143 | 2 | 89 | MEF2 CHIMERA |
| 6bz1_D | 2e-20 | 56 | 143 | 2 | 89 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the development of floral organs. Acts as a C-class protein in association with MADS3. Involved in the control of lodicule number (whorl 2), stamen specification (whorl 3), floral meristem determinacy and regulation of the carpel morphogenesis (whorl 4). Plays a more predominant role in floral meristem determinacy than MADS3. {ECO:0000269|PubMed:16326928}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
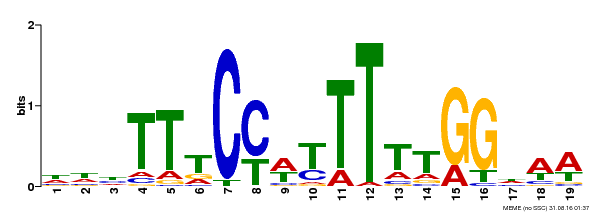
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do004107.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | MZEZAG1A | 0.0 | L18924.1 Corn ZAG1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004960599.1 | 0.0 | MADS-box transcription factor 58 | ||||
| Swissprot | Q2V0P1 | 1e-132 | MAD58_ORYSJ; MADS-box transcription factor 58 | ||||
| TrEMBL | A0A1E5VH85 | 0.0 | A0A1E5VH85_9POAL; MADS-box transcription factor 58 (Fragment) | ||||
| STRING | Pavir.Ca00579.1.p | 1e-178 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP649 | 37 | 144 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 4e-95 | MIKC_MADS family protein | ||||



