 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do011504.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 648aa MW: 67579.9 Da PI: 7.1407 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 38.1 | 3.7e-12 | 301 | 357 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng.krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++gr++A+++d ++ g rk + g ++ +e+Aa+a++ a++k++g
Do011504.1 301 SQYRGVTRHRWTGRYEAHLWDnSCKkEGqTRKGRQ-GGYDMEEKAARAYDLAALKYWG 357
78*******************77776664346655.77******************98 PP
| |||||||
| 2 | AP2 | 47.9 | 3.3e-15 | 400 | 451 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s y+GV+++++ grW A+I +s +k +lg+f t eeAa+a++ a+ k++g
Do011504.1 400 SIYRGVTRHHQHGRWQARIGRVSG---NKDLYLGTFSTQEEAAEAYDVAAIKFRG 451
57****************999754...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 2.6E-9 | 301 | 357 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.47E-14 | 301 | 366 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 1.69E-17 | 301 | 366 | No hit | No description |
| SMART | SM00380 | 4.4E-22 | 302 | 371 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.293 | 302 | 365 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.4E-12 | 302 | 365 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.8E-6 | 303 | 314 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 1.7E-10 | 400 | 451 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 7.85E-18 | 400 | 460 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 6.63E-24 | 400 | 461 | No hit | No description |
| PROSITE profile | PS51032 | 19.137 | 401 | 459 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.9E-34 | 401 | 465 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 1.9E-18 | 401 | 459 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.8E-6 | 441 | 461 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007276 | Biological Process | gamete generation | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 648 aa Download sequence Send to blast |
MTSNSCQNLS SCSTGGSDAA VGGGGSWLGF SLSPHMAGTI DGGDGSNAVQ VQQHHGGLFY 60 PPVVSSSPAP FCYALGGGQD GVATAGANGG GVFYPGLSSM PLKADGSLCI MEVLHRSEQE 120 HHGVVVSSAS PKLEDFLGAG PTMALSLDNS SFYYGGGHGH HGHGQDQAAY LQPLHCAVIP 180 GSGGHGHDVY GGHAQLVDEQ SAAAMAASWF SARGGGGYDV NGAGAILPVQ GHAHPLSLSM 240 SSGTGSQSSC VTMQVGPYPH ADAVAEYIAM DGSKKRGGGG AGQKQPTVHR KSIDTFGQRT 300 SQYRGVTRHR WTGRYEAHLW DNSCKKEGQT RKGRQGGYDM EEKAARAYDL AALKYWGTST 360 HINFPIEDYR EELEEMKNMT RQEYVAHLRR KSSGFSRGAS IYRGVTRHHQ HGRWQARIGR 420 VSGNKDLYLG TFSTQEEAAE AYDVAAIKFR GLNAVTNFDI TRYDVDKIME SNTLLPGEQV 480 RRKKDGEGGV SESDAVTNAA AALVQAGNCA AADTWKIQGA AAALPAVARS DALQVHGGQQ 540 HQDLLSSEAF SLLHGIVSVD AGGQGGSASA QMSNASSLAP SVSNSREQSP DRGVGGGGLA 600 MLFAKPAAAA SKLACPLPLG SWVSPSPVSA RPGVSIAHLP MFAAWTDA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Acts as positive regulator of adventitious (crown) root formation by promoting its initiation. Promotes adventitious root initiation through repression of cytokinin signaling by positively regulating the two-component response regulator RR1. Regulated by the auxin response factor and transcriptional activator ARF23/ARF1. {ECO:0000269|PubMed:21481033}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00088 | SELEX | Transfer from AT4G37750 | Download |
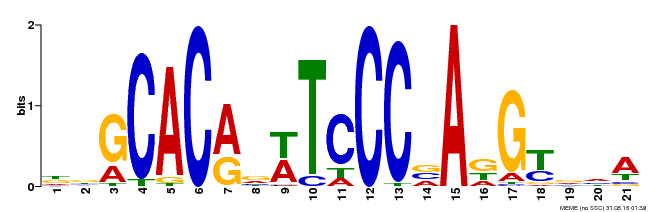
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do011504.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by auxin. {ECO:0000269|PubMed:21481033}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375318 | 1e-176 | AK375318.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3090N20. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004981612.1 | 0.0 | AP2-like ethylene-responsive transcription factor ANT isoform X4 | ||||
| Swissprot | Q84Z02 | 1e-169 | CRL5_ORYSJ; AP2-like ethylene-responsive transcription factor CRL5 | ||||
| TrEMBL | A0A1E5VRG8 | 0.0 | A0A1E5VRG8_9POAL; AP2-like ethylene-responsive transcription factor ANT | ||||
| STRING | Pavir.J24038.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP948 | 38 | 133 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37750.1 | 1e-122 | AP2 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



