 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do012100.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 326aa MW: 34073.1 Da PI: 7.8386 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.7 | 1.1e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+ lv +v+++G+g+W++++ g+ R+ k+c++rw +yl
Do012100.1 14 KGPWTPEEDLMLVSYVQEHGPGNWRAVPTNTGLMRCSKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 51.3 | 2.8e-16 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T +E++l+v++ ++lG++ W++Ia++++ Rt++++k++w+++l
Do012100.1 67 RGNFTDQEEKLIVHLQALLGNR-WAAIASYLP-ERTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-24 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 24.935 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.13E-30 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.5E-14 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.92E-11 | 16 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-24 | 65 | 117 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.427 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.3E-16 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.5E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.62E-11 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0080167 | Biological Process | response to karrikin | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MGRPPCCDKV GVKKGPWTPE EDLMLVSYVQ EHGPGNWRAV PTNTGLMRCS KSCRLRWTNY 60 LRPGIKRGNF TDQEEKLIVH LQALLGNRWA AIASYLPERT DNDIKNYWNT HLKKKLKKMQ 120 AGEGDGGDDG AGASEGGDAG GRAGAGGGKR PAIPKGQWER RLQTDIHTAR QALRDALSLE 180 PSAPPANKVA PPPMTPPGST TYASSAENIA RLLEGWLRPG GGSGGKGTEA SGSTSTTATT 240 QQRPQCSGEG AASASASHSS GAAGNMAAQT PECSTETSKM AGSAAGAGSA PAFSMLESWL 300 LDDGMGHGEV GLMADVVPLG DPSEFF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 1e-26 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 1e-26 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 139 | 147 | GGRAGAGGG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00395 | DAP | Transfer from AT3G47600 | Download |
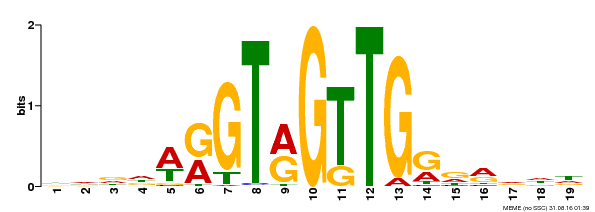
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do012100.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC165178 | 0.0 | AC165178.2 Zea mays clone ZMMBBb-272P17, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008652703.1 | 0.0 | myb-related protein 306 | ||||
| Swissprot | P81392 | 1e-108 | MYB06_ANTMA; Myb-related protein 306 | ||||
| TrEMBL | A0A096QJ49 | 0.0 | A0A096QJ49_MAIZE; Transcription factor MYB30 | ||||
| TrEMBL | A0A3L6DZ53 | 0.0 | A0A3L6DZ53_MAIZE; Myb-related protein 306 | ||||
| STRING | GRMZM2G045748_P02 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP229 | 38 | 296 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G47600.1 | 6e-98 | myb domain protein 94 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



