 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do016288.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 412aa MW: 45441.9 Da PI: 8.5913 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.5 | 1.1e-05 | 61 | 83 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C++C+k F r nL+ H r H
Do016288.1 61 FVCEICNKGFQRDQNLQLHRRGH 83
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13.9 | 0.00016 | 147 | 169 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cgk++ +s+ k H + +
Do016288.1 147 WRCERCGKRYAVQSDWKAHVKGC 169
58*****************9866 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-5 | 60 | 83 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 9.98E-9 | 60 | 83 | No hit | No description |
| SMART | SM00355 | 0.017 | 61 | 83 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF12171 | 6.3E-5 | 61 | 83 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.949 | 61 | 83 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 63 | 83 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 89 | 112 | 142 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 9.3E-5 | 135 | 167 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 9.98E-9 | 142 | 167 | No hit | No description |
| SMART | SM00355 | 95 | 147 | 167 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 412 aa Download sequence Send to blast |
MRAPFLHQEV FHLLWLRLVS LILVGWLSAL TTPMDATRET KDPGAEVIAL SPRTLVATNR 60 FVCEICNKGF QRDQNLQLHR RGHNLPWKLR QRSLPSGAGR QGDGAPPRKR VYVCPEPTCV 120 HHDPARALGD LTGIKKHFSR KHGEKRWRCE RCGKRYAVQS DWKAHVKGCG TREYRCDCGI 180 LFTRYNTYCS CVLVCMHGAM DTLISHEQAL PDQEPRSCPG LLGFLLFLKD SLLTHRAFCD 240 ALAEESARLL AAANNSSTIT TNSSNGNSSS ITTPLFLPFP NNPPLAAAQN PNAIYFLHQE 300 QAPPFLQPRM MQPSPYLDLH AGATFTTAGG GIVTDTVNFG LAPDGSVAMR AAGGGHRRLT 360 RDFLGVDGGG QVEELQLQLY AATAAASCAT DLTRQYLGER LPPVSETWSH NF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 3e-26 | 143 | 256 | 3 | 72 | Zinc finger protein JACKDAW |
| 5b3h_F | 3e-26 | 143 | 256 | 3 | 72 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that acts as a flowering master switch in both long and short days, independently of the circadian clock (By similarity). Promotes flowering upstream of HD1 by up-regulating FTL1, FTL4, FTL5, FTL6, EHD1, HD3A and RFT1 (By similarity). Seems to repress FTL11 expression (By similarity). May recognize the consensus motif 5'-TTTGTCGTAAT-3' in target gene promoters (By similarity). {ECO:0000250|UniProtKB:B1B534}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00673 | SELEX | Transfer from GRMZM2G011357 | Download |
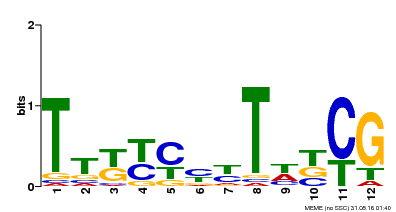
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do016288.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK361456 | 1e-132 | AK361456.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1141C05. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004983079.1 | 1e-161 | protein indeterminate-domain 12 | ||||
| Swissprot | B8BGV5 | 1e-124 | EHD2_ORYSI; Protein EARLY HEADING DATE 2 | ||||
| TrEMBL | A0A1E5VCE4 | 0.0 | A0A1E5VCE4_9POAL; Protein indeterminate-domain 2 | ||||
| STRING | Si036003m | 1e-160 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP13096 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G50700.1 | 8e-79 | indeterminate(ID)-domain 2 | ||||



