 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Do016545.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Dichantheliinae; Dichanthelium
|
||||||||
| Family | GRAS | ||||||||
| Protein Properties | Length: 618aa MW: 65197 Da PI: 4.889 | ||||||||
| Description | GRAS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GRAS | 465.1 | 4.1e-142 | 236 | 614 | 1 | 374 |
GRAS 1 lvelLlecAeavssgdlelaqalLarlselaspdgdpmqRlaayfteALaarlarsvselykalppsetseknsseelaalkl...fsevsPilkfshl 96
lv++Ll+cAeav+++++++a+al++++ las++g +m+++aayf eALa+r++r l+p ++s+ ++++a+ l f+e +P+lkf+h+
Do016545.1 236 LVHALLACAEAVQQENFTAADALVKQIPMLASSQGGAMRKVAAYFGEALARRVYR--------LRPAQDSS--LLDAAFADLLhahFYESCPYLKFAHF 324
689****************************************************........55555553..4444444444446************* PP
GRAS 97 taNqaIleavegeervHiiDfdisqGlQWpaLlqaLasRpegppslRiTgvgspesgskeeleetgerLakfAeelgvpfefnvlvakrledleleeLr 195
taNqaIlea++g++rvH++Df+i+qG+QWpaLlqaLa Rp+gpps+R+Tgvg+p+++++++l+++g++La+fA++++v+f+++ lva +l+dle+ +L+
Do016545.1 325 TANQAILEAFAGCRRVHVVDFGIKQGMQWPALLQALALRPGGPPSFRLTGVGPPQPDETDALQQVGWKLAQFAHTIRVDFQYRGLVAATLADLEPFMLQ 423
**************************************************************************************************5 PP
GRAS 196 vkp......gEalaVnlvlqlhrlldesvsleserdevLklvkslsPkvvvvveqeadhnsesFlerflealeyysalfdsleak........lpres. 279
+ E++aVn+v++lhrll+++++le+ vL +v+ ++P++v+vveqea+hns+sF++rf+e+l+yys++fdsle p+++
Do016545.1 424 PEGedtddePEVIAVNSVFELHRLLAQPGALEK----VLGTVRAVRPRIVTVVEQEANHNSGSFVDRFTESLHYYSTMFDSLEGAgggqsadaSPAAAg 518
4444445579***********************....*********************************************87767777775444447 PP
GRAS 280 eerikvErellgreivnvvacegaerrerhetlekWrerleeaGFkpvplsekaakqaklllrkvk.sdgyrveeesgslvlgWkdrpLvsvSaWr 374
+++++++++lgr+i+nvvacegaer+erhetl++Wr+rl++aGF+pv+l+++a+kqa++ll+ ++ +dgyrvee++g+l+lgW++rpL+++SaWr
Do016545.1 519 ATDQVMSEVYLGRQICNVVACEGAERTERHETLGQWRNRLGRAGFEPVHLGSNAYKQASTLLALFAgGDGYRVEEKDGCLTLGWHTRPLIATSAWR 614
999********************************************************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF12041 | 2.4E-36 | 34 | 107 | IPR021914 | Transcriptional factor DELLA, N-terminal |
| SMART | SM01129 | 6.2E-33 | 34 | 113 | No hit | No description |
| PROSITE profile | PS50985 | 68.47 | 210 | 593 | IPR005202 | Transcription factor GRAS |
| Pfam | PF03514 | 1.4E-139 | 236 | 614 | IPR005202 | Transcription factor GRAS |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042176 | Biological Process | regulation of protein catabolic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:2000033 | Biological Process | regulation of seed dormancy process | ||||
| GO:2000377 | Biological Process | regulation of reactive oxygen species metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MKREYQDAGG SGGDMGSSKD KMMAAAGGQE EEVDELLAAL GYKVRSSDMA DVAQKLEQLE 60 MAMGMGSTAA AADDGFVSHL ATDTVHYNPS DLSSWVESML SELNAPPPPL SPAPPAPRLA 120 STSSTVTGGG AAAGGSYFDL PPAVDSSSST YALKPIPSPV TASADPSTDS TREPKRMRTG 180 GGSTSSSSSS SSSLDGGRTR SSVVEAAPPA TQASAAANGP AVPVVVVDTQ DAGIRLVHAL 240 LACAEAVQQE NFTAADALVK QIPMLASSQG GAMRKVAAYF GEALARRVYR LRPAQDSSLL 300 DAAFADLLHA HFYESCPYLK FAHFTANQAI LEAFAGCRRV HVVDFGIKQG MQWPALLQAL 360 ALRPGGPPSF RLTGVGPPQP DETDALQQVG WKLAQFAHTI RVDFQYRGLV AATLADLEPF 420 MLQPEGEDTD DEPEVIAVNS VFELHRLLAQ PGALEKVLGT VRAVRPRIVT VVEQEANHNS 480 GSFVDRFTES LHYYSTMFDS LEGAGGGQSA DASPAAAGAT DQVMSEVYLG RQICNVVACE 540 GAERTERHET LGQWRNRLGR AGFEPVHLGS NAYKQASTLL ALFAGGDGYR VEEKDGCLTL 600 GWHTRPLIAT SAWRVAAP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_A | 4e-61 | 243 | 613 | 25 | 377 | Protein SCARECROW |
| 5b3h_D | 4e-61 | 243 | 613 | 25 | 377 | Protein SCARECROW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional regulator that acts as a repressor of the gibberellin (GA) signaling pathway. Probably acts by participating in large multiprotein complexes that repress transcription of GA-inducible genes. Upon GA application, it is degraded by the proteasome, allowing the GA signaling pathway. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00611 | ChIP-seq | Transfer from AT2G01570 | Download |
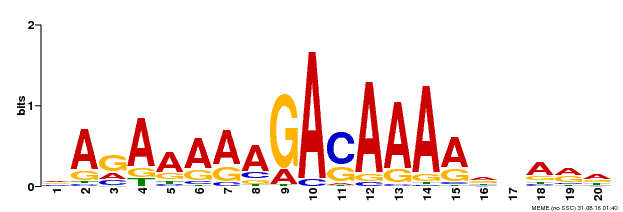
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Do016545.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ242530 | 0.0 | AJ242530.1 Zea mays partial d8 gene for gibberellin response modulator. | |||
| GenBank | BT054262 | 0.0 | BT054262.1 Zea mays full-length cDNA clone ZM_BFb0203D23 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001354393.1 | 0.0 | DELLA protein DWARF8 | ||||
| Swissprot | Q9ST48 | 0.0 | DWRF8_MAIZE; DELLA protein DWARF8 | ||||
| TrEMBL | A0A1E5VC25 | 0.0 | A0A1E5VC25_9POAL; DELLA protein DWARF8 | ||||
| STRING | GRMZM2G144744_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP998 | 38 | 138 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14920.1 | 0.0 | GRAS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



